7M14
 
 | | x-ray structure of cj1430 in the presence of GDP, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M13
 
 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
4MZ1
 
 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12 | | Descriptor: | 1-(4-bromophenyl)-3-{2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}urea, ACETIC ACID, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-28 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3991 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase, with a Internal Deletion of CBS Domain from Campylobacter jejuni complexed with inhibitor compound P12
To be Published, 2013
|
|
7Z23
 
 | | Connexin43 hemi channel in nanodisc | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Qi, C, Korkhov, M.V. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-08 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structure of the connexin-43 gap junction channel in a putative closed state.
Elife, 12, 2023
|
|
2LRK
 
 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose | | Descriptor: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, Phosphocarrier protein HPr | | Authors: | Cai, M, Jung, Y, Clore, M. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
7C65
 
 | | Crystal structure of thioredoxin m1 | | Descriptor: | SODIUM ION, Thioredoxin M1, chloroplastic | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C2B
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin f2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7949 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
7C3F
 
 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin m2 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, Ferredoxin-thioredoxin reductase variable chain, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3986 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
2X63
 
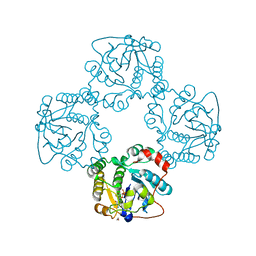 | | Crystal structure of the sialyltransferase CST-II N51A in complex with CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
2X62
 
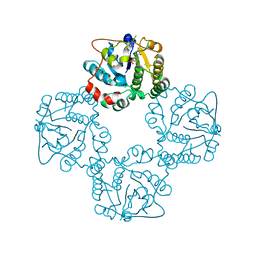 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE CST-II Y81F IN COMPLEX WITH CMP | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-2,3-/2,8-SIALYLTRANSFERASE, N3-PROTONATED CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
7BZK
 
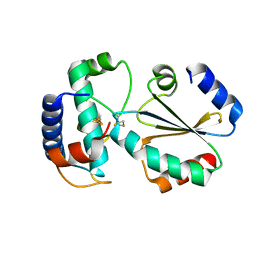 | | Crystal structure of ferredoxin: thioredoxin reductase and thioredoxin y1 complex | | Descriptor: | Ferredoxin-thioredoxin reductase catalytic chain, chloroplastic, IRON/SULFUR CLUSTER, ... | | Authors: | Kurisu, G, Juniar, L, Tanaka, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5935 Å) | | Cite: | Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Protein Sci., 29, 2020
|
|
4XAK
 
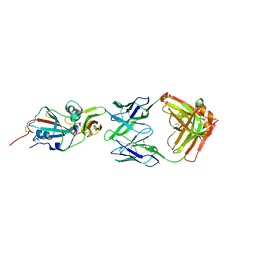 | | Crystal structure of potent neutralizing antibody m336 in complex with MERS Co-V RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy chain of neutralizing antibody m336, ... | | Authors: | Zhou, T, Dimtrov, D.S, Ying, T. | | Deposit date: | 2014-12-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody.
Nat Commun, 6, 2015
|
|
2X1I
 
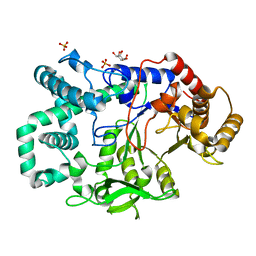 | | glycoside hydrolase family 77 4-alpha-glucanotransferase from thermus brockianus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 4-ALPHA-GLUCANOTRANSFERASE, PHOSPHATE ION, ... | | Authors: | Yoon, S.-M, Jung, J.-H, Jung, T.-Y, Song, H.-N, Park, C.-S, Woo, E.-J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural and Functional Analysis of Substrate Recognition by the 250S Loop in Amylomaltase from Thermus Brockianus.
Proteins, 79, 2011
|
|
398D
 
 | | 3'-DNA-RNA-5' JUNCTION FORMED DURING INITIATION OF MINUS-STRAND SYNTHESIS OF HIV REPLICATION | | Descriptor: | DNA/RNA (5'-R(*GP*CP*CP*AP)-D(*CP*TP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*UP*GP*GP*C)-3') | | Authors: | Mueller, U, Meier, G, Mochi-Onori, A, Cellai, L, Heumann, H. | | Deposit date: | 1998-05-04 | | Release date: | 1998-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of an eight-base pair duplex containing the 3'-DNA-RNA-5' junction formed during initiation of minus-strand synthesis of HIV replication.
Biochemistry, 37, 1998
|
|
6B6G
 
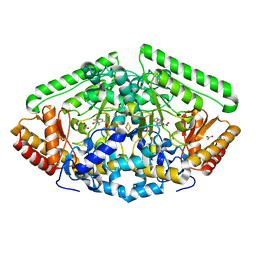 | | Crystal Structure of GABA Aminotransferase bound to (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic acid, an Potent Inactivatorfor the Treatment of Addiction | | Descriptor: | (3R,4E)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]cyclopent-1-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Mascarenhas, R, Juncosa, J.I, Takaya, K, Le, L.V, Moschitto, M.J, Silverman, R.B, Liu, D. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Mechanism of (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic Acid, a Highly Potent gamma-Aminobutyric Acid Aminotransferase Inactivator for the Treatment of Addiction.
J. Am. Chem. Soc., 140, 2018
|
|
3QB7
 
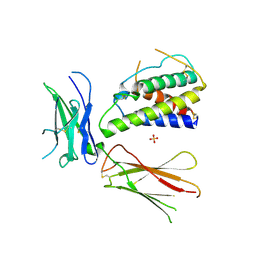 | | Interleukin-4 mutant RGA bound to cytokine receptor common gamma | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Bates, D.L, Junttila, I.S, Creusot, R.J, Moraga, I, Lupardus, P, Fathman, C.G, Paul, W.E, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines.
Nat.Chem.Biol., 8, 2012
|
|
7LAM
 
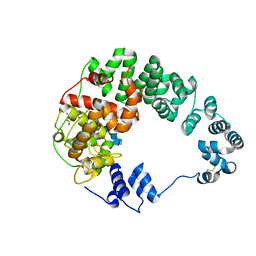 | | Crystal structure of Campylobacter jejuni Cj0843c lytic transglycosylase in complex with N,N',N''-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CITRIC ACID, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Turnover Chemistry and Structural Characterization of the Cj0843c Lytic Transglycosylase of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7LAQ
 
 | |
1L6B
 
 | |
3GET
 
 | |
2NQD
 
 | | Crystal structure of cysteine protease inhibitor, chagasin, in complex with human cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin L, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-31 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
2NNR
 
 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | Authors: | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
1IXS
 
 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
