3PC6
 
 | |
1QJB
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
2DXC
 
 | | Recombinant thiocyanate hydrolase, fully-matured form | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Arakawa, T, Kawano, Y, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2006-08-25 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Catalytic Activation of Thiocyanate Hydrolase Involving Metal-Ligated Cysteine Modification
J.Am.Chem.Soc., 131, 2009
|
|
3V7D
 
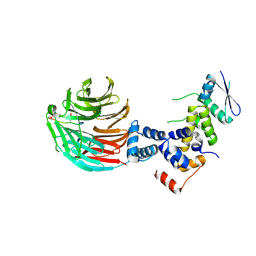 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
6OVP
 
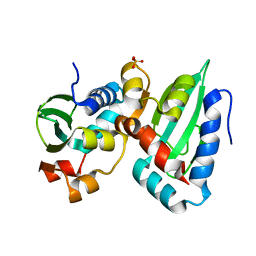 | |
9IXD
 
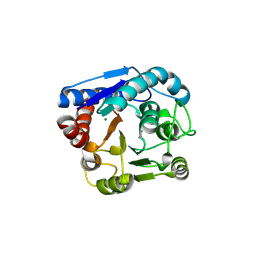 | |
9IXF
 
 | |
9IXE
 
 | |
4NPP
 
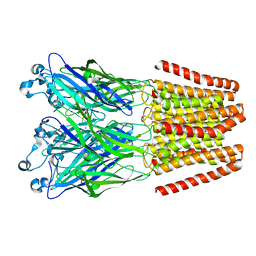 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9IXC
 
 | |
9IXG
 
 | |
3HB8
 
 | | Structures of Thymidylate Synthase R163K with Substrates and Inhibitors Show Subunit Asymmetry | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Gibson, L.M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2EG8
 
 | | The crystal structure of E. coli dihydroorotase complexed with 5-fluoroorotic acid | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-02-28 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design
J.Mol.Biol., 370, 2007
|
|
3PLY
 
 | | Structure of Oxidized P96G Mutant of Amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2010-11-15 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline 96 of the copper ligand loop of amicyanin regulates electron transfer from methylamine dehydrogenase by positioning other residues at the protein-protein interface.
Biochemistry, 50, 2011
|
|
6H7Y
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor RNA 1-79-1 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(4-fluorophenyl)amino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2018-07-31 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
6QIR
 
 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6U30
 
 | | The crystal structure of 4-pyridin-3-ylbenzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-3-yl)benzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
3H9K
 
 | | Structures of Thymidylate Synthase R163K with Substrates and Inhibitors Show Subunit Asymmetry | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Gibson, L.M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3VHB
 
 | | IMIDAZOLE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | Descriptor: | IMIDAZOLE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | Deposit date: | 1999-03-17 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
4YU0
 
 | | Crystal structure of a tetramer of GluA2 TR mutant ligand binding domains bound with glutamate at 1.26 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Chebli, M, Salazar, H, Baranovic, J, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the tetrameric GluA2 ligand-binding domain in complex with glutamate at 1.26 Angstroms resolution
To Be Published
|
|
7BLC
 
 | | BAZ2A bromodomain in complex with compound UP39 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, methyl 5-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]-2-(2-piperazin-1-ylethoxy)benzoate | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
7BL8
 
 | | BAZ2A bromodomain in complex with the chemical probe BAZ2-ICR | | Descriptor: | 4-[5-(1-methylpyrazol-4-yl)-3-[2-(1-methylpyrazol-4-yl)ethyl]imidazol-4-yl]benzenecarbonitrile, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reevaluation of bromodomain ligands targeting BAZ2A.
Protein Sci., 32, 2023
|
|
