7YJ0
 
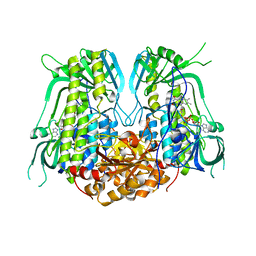 | |
7YIT
 
 | |
7YIA
 
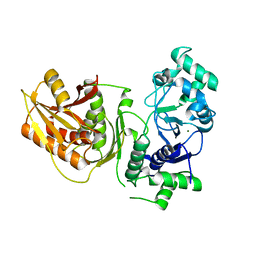 | | Crystal structure of K74A mutant of Cap4 SAVED domain-containing receptor from Enterobacter cloacae | | Descriptor: | CD-NTase-associated protein 4, MAGNESIUM ION | | Authors: | Ko, T.-P, Yang, C.-S, Hou, M.-H, Wang, Y.-C, Chen, Y. | | Deposit date: | 2022-07-15 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Specific recognition of cyclic oligonucleotides by Cap4 for phage infection.
Int.J.Biol.Macromol., 237, 2023
|
|
7YI5
 
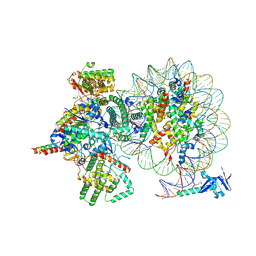 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI4
 
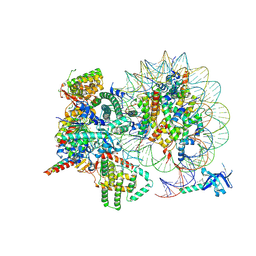 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI3
 
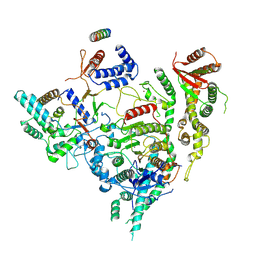 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI2
 
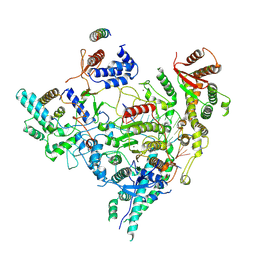 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
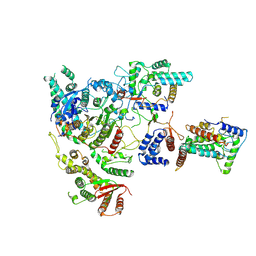 | | Cryo-EM structure of Rpd3S complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
7YHJ
 
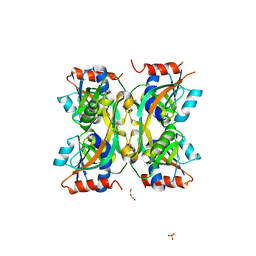 | |
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH4
 
 | |
7YH3
 
 | |
7YH2
 
 | | TRAPPC3 from Thorarchaeota AB25 | | Descriptor: | TRAPPC3 from Thorarchaeota AB25, ZINC ION | | Authors: | Robinson, R.C, Tran, L.T. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | TRAPPC3 from Thorarchaeota AB25
To Be Published
|
|
7YH1
 
 | |
7YGW
 
 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGV
 
 | | Crystal structure of the Ca2+-bound EFhd1/Swiprosin-2 | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Ynag, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGQ
 
 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7YGP
 
 | | DNA duplex containing 5OHU-T base pairs | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*AP*CP*CP*TP*(OHU)P*GP*GP*TP*CP*C)-3') | | Authors: | Kondo, J, Torigoe, H, Arakawa, F. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Specific binding of Hg 2+ to mismatched base pairs involving 5-hydroxyuracil in duplex DNA.
J.Inorg.Biochem., 241, 2023
|
|
7YGO
 
 | | DNA duplex containing 5OHU-Hg(II)-T base pairs | | Descriptor: | DNA (5'-D(*GP*GP*AP*CP*CP*TP*(6HU)P*GP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Kondo, J, Torigoe, H, Arakawa, F. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Specific binding of Hg 2+ to mismatched base pairs involving 5-hydroxyuracil in duplex DNA.
J.Inorg.Biochem., 241, 2023
|
|
7YGF
 
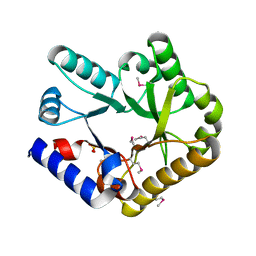 | | Crystal structure of YggS from Fusobacterium nucleatum | | Descriptor: | Pyridoxal phosphate homeostasis protein, SULFATE ION | | Authors: | He, S.R, Chan, Y.Y, Wang, L.L, Bai, X, Bu, T.T, Zhang, J, Xu, Y.B. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum.
Molecules, 27, 2022
|
|
7YFT
 
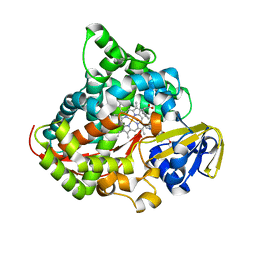 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82C/L181M in complex with N-imidazolyl-pentanoyl-L-phenylalanine, indane and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, 2,3-dihydro-1H-indene, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YFS
 
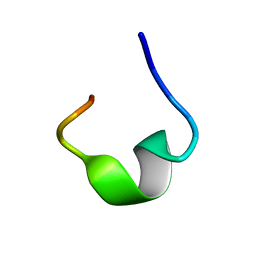 | | The NMR structure of noursin, a tricyclic ribosomal peptide containing a histidine-to-butyrine crosslink | | Descriptor: | noursin | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
7YFP
 
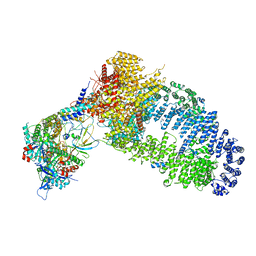 | | The NuA4 histone acetyltransferase complex from S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
