5T6H
 
 | |
4QLJ
 
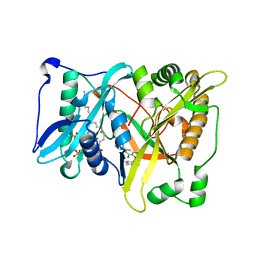
 | | Crystal structure of rice BGlu1 E386G/Y341A/Q187A mutant complexed with cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2014-06-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Effects of active site cleft residues on oligosaccharide binding, hydrolysis, and glycosynthase activities of rice BGlu1 and its mutants
Protein Sci., 23, 2014
|
|
7E9S
 
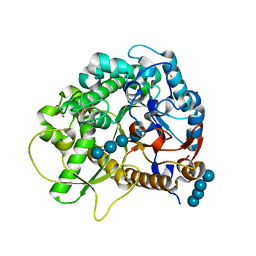
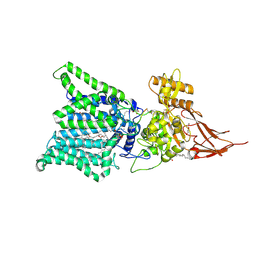 | | Archaeal oligosaccharyltransferase AglB from Archaeoglobus fulgidus in complex with an inhibitory peptide and a dolichol-phosphate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, DI(HYDROXYETHYL)ETHER, Dolichyl-phosphooligosaccharide-protein glycotransferase 3, ... | | Authors: | Taguchi, Y, Hirata, K, Kohda, D. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Commun Biol, 4, 2021
|
|
7EBL
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), STING | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7EBD
 
 | | Bacterial STING in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, TIR-like domain-containing protein | | Authors: | Ko, T.-P, Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and functional implication of bacterial STING.
Nat Commun, 13, 2022
|
|
7MYB
 
 | |
6S9Z
 
 | | Expression tag modified N-terminus of human Carbonic Anhydrase II covalently linked to fragment | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3-[3,4-bis(fluoranyl)phenyl]-5-methyl-1$l^{4},4,5$l^{4},7-tetrazabicyclo[4.3.0]nona-1(6),2,4-triene, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-07-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | A Proof-of-Concept Fragment Screening of a Hit-Validated 96-Compounds Library against Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
6PC5
 
 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8OV5
 
 | |
5F9T
 
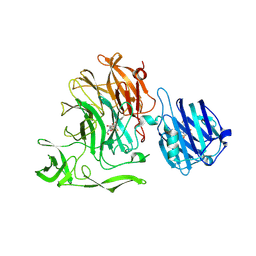 | | Crystal Structure of Streptococcus pneumoniae NanC, covalent complex with a fluorinated Neu5Ac derivative | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-12-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
5U24
 
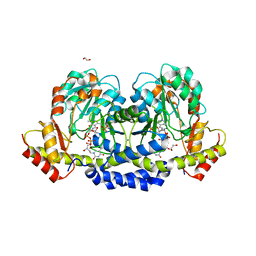 | | X-ray structure of the WlaRG aminotransferase from campylobacter jejuni, K184A mutant in complex with TDP-Fuc3N | | Descriptor: | (2R,3R,4S,5R,6R)-3,5-dihydroxy-4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
7ZP7
 
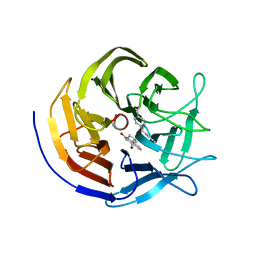 | | Crystal structure of evolved photoenzyme EnT1.3 (truncated) with bound product | | Descriptor: | (1~{R},10~{R},12~{S})-15-oxa-8-azatetracyclo[8.5.0.0^{1,12}.0^{2,7}]pentadeca-2(7),3,5-trien-9-one, 1,2-ETHANEDIOL, EnT1.3 C | | Authors: | Hardy, F.J, Levy, C. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A designed photoenzyme for enantioselective [2+2] cycloadditions.
Nature, 611, 2022
|
|
5U21
 
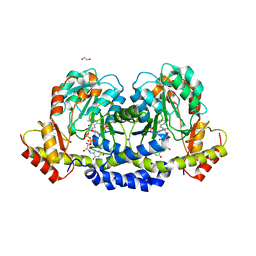 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
7E4P
 
 | | Crystal structure of tubulin in complex with Ansamitocin P3 | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C3 Ester Side Chain Plays a Pivotal Role in the Antitumor Activity of Maytansinoids
To Be Published
|
|
7E4Y
 
 | | Crystal structure of tubulin in complex with L-DM4-SMe | | Descriptor: | (1S,2S,3S,5R,6S,16E,18E,20S,21R)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-methyl-3-(methyldisulfanyl)butanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | C3 Ester Side Chain Plays a Pivotal Role in the Antitumor Activity of Maytansinoids
To Be Published
|
|
7E5Z
 
 | | Dehydrogenase holoenzyme | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Roh, S.H, Park, J.S, Heo, Y.Y. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dehydrogenase holoenzyme
To Be Published
|
|
8FR3
 
 | | E. coli EF-Tu in complex with KKL-55 | | Descriptor: | 3-chloro-N-(1-propyl-1H-tetrazol-5-yl)benzamide, Elongation factor Tu, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nguyen, H.A, Kuzmishin Nagy, A.B, Dunham, C.M. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Antibiotic that inhibits trans -translation blocks binding of EF-Tu to tmRNA but not to tRNA.
Mbio, 14, 2023
|
|
6GWE
 
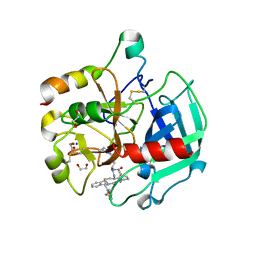 | | Crystal structure of Thrombin bound to P2 macrocycle | | Descriptor: | (10S,14S,17R)-14-(3-carbamimidamidopropyl)-3-[[2-(hydroxymethyl)phenyl]methyl]-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(21),22,24-triene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cendron, L, Angelini, A, Kale, S.S, Bergeron-Brlek, M, Wu, Y, Heinis, C. | | Deposit date: | 2018-06-23 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thiol-to-amine cyclization reaction enables screening of large libraries of macrocyclic compounds and the generation of sub-kilodalton ligands.
Sci Adv, 5, 2019
|
|
5IP6
 
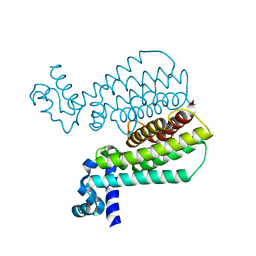 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with N-((tetrahydrofuran-3-yl)methyl)pyrrolidine-1-carboxamide at 1.93A resolution | | Descriptor: | N-{[(3S)-oxolan-3-yl]methyl}pyrrolidine-1-carboxamide, TetR-family transcriptional regulatory repressor protein | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
6V8Q
 
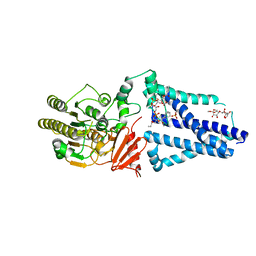 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
6EVQ
 
 | |
6UXE
 
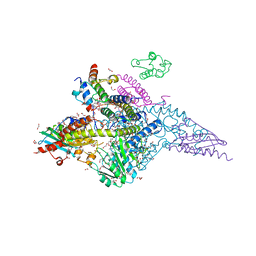 | | Structure of the human mitochondrial desulfurase complex Nfs1-ISCU2(M140I)-ISD11 with E.coli ACP1 at 1.57 A resolution showing flexibility of N terminal end of ISCU2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2019-11-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6PC6
 
 | | E. coli 50S ribosome bound to compound 47 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
4I5T
 
 | | Crystal structure of yeast Ap4A phosphorylase Apa2 | | Descriptor: | 5',5'''-P-1,P-4-tetraphosphate phosphorylase 2 | | Authors: | Jiang, Y.L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of yeast Apa2 reveal catalytic insights into a canonical AP4A phosphorylase of the histidine triad superfamily
J.Mol.Biol., 425, 2013
|
|
1FO6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE | | Descriptor: | N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE, XENON | | Authors: | Wang, W.-C, Hsu, W.-H, Chien, F.-T, Chen, C.-Y. | | Deposit date: | 2000-08-25 | | Release date: | 2001-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft.
J.Mol.Biol., 306, 2001
|
|
