4X6T
 
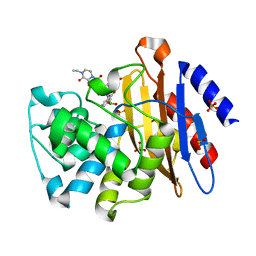 | | M.tuberculosis betalactamase complexed with inhibitor EC19 | | Descriptor: | 3-[(2R)-2-(dihydroxyboranyl)-2-{[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibiting the beta-Lactamase of Mycobacterium tuberculosis (Mtb) with Novel Boronic Acid Transition-State Inhibitors (BATSIs).
ACS Infect Dis, 1, 2015
|
|
6OAL
 
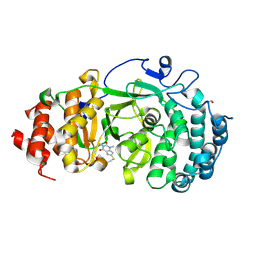 | | Structure of human PARG complexed with JA2120 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Ahmed, Z, Warden, S, Tainer, J.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
7Q07
 
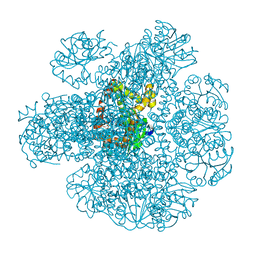 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the open state with NADP and tartrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ketol-Acid Reductoisomerase from Methanothermococcus thermolithotrophicus, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
4WKQ
 
 | |
7Q03
 
 | | Ketol-acid reductoisomerase from Methanothermococcus thermolithotrophicus in the close state with NADP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, GLYCEROL, ... | | Authors: | Lemaire, O.N, Mueller, M, Wagner, T. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen.
Biomolecules, 11, 2021
|
|
4WRG
 
 | |
1EN8
 
 | |
6NT0
 
 | | Catalase 3 from N.Crassa in ferrous state, X-ray reduced (1.315 MGy) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Catalase-3, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E, Stojanoff, V. | | Deposit date: | 2019-01-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
6PBL
 
 | |
9VKF
 
 | |
9NT0
 
 | | OXA-23-meropenem, pH 7.5 | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase OXA-23, SULFATE ION | | Authors: | Smith, C.A, Toth, M, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-03-17 | | Release date: | 2025-08-06 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dual mechanism of the OXA-23 carbapenemase inhibition by the carbapenem NA-1-157.
Antimicrob.Agents Chemother., 2025
|
|
6O4H
 
 | | Structure of ALDH7A1 mutant A171V complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
4V2L
 
 | |
4V2M
 
 | |
7P9E
 
 | |
7PJM
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
9MLT
 
 | | Crystal structure of dihydrofolate reductase (DHFR) from the filarial nematode W. bancrofti in complex with NADPH and [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-cyanophenyl]acetic acid (TSD25 or OFD) | | Descriptor: | Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-cyanophenyl]acetic acid | | Authors: | Frey, K.M, Goodey, N.M, Kwarteng, S, Wilhelm, J. | | Deposit date: | 2024-12-19 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | A virtual screening strategy to repurpose antifolate compounds as W.bancrofti DHFR inhibitors.
Bioorg.Med.Chem.Lett., 129, 2025
|
|
6VJ4
 
 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | Descriptor: | Peptidylprolyl isomerase PrsA | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
8F7Z
 
 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
6O4C
 
 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
4WB9
 
 | | Human ALDH1A1 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
6O4B
 
 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4F
 
 | | Structure of ALDH7A1 mutant N167S complexed with alpha-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6YXF
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6P0E
 
 | | Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
