4Q9Q
 
 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4N6U
 
 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4N6T
 
 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4Q9R
 
 | | Crystal structure of an RNA aptamer bound to trifluoroethyl-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
6MTM
 
 | | Crystal Structure of EM2 TCR in complex with HLA-B*37:01-NP338 | | Descriptor: | Beta-2-microglobulin, EM2 TCR alpha chain, EM2 TCR beta chain, ... | | Authors: | Gras, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Broad CD8+T cell cross-recognition of distinct influenza A strains in humans.
Nat Commun, 9, 2018
|
|
6J36
 
 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
6D4M
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
7OLZ
 
 | | Crystal structure of the SARS-CoV-2 RBD with neutralizing-VHHs Re5D06 and Re9F06 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Nanobody Re5D06, Nanobody Re9F06, ... | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
7ON5
 
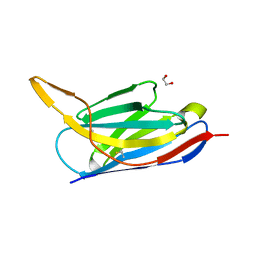 | | Crystal structure of the SARS-CoV-2 neutralizing nanobody Re5D06 | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Nanobody Re5D06 | | Authors: | Aksu, M, Guttler, T, Gorlich, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies.
Embo J., 40, 2021
|
|
6D4E
 
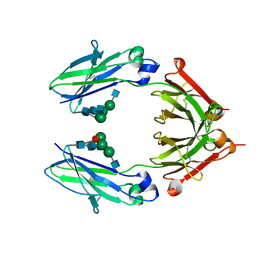 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Fc Fragment of IgG1 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D4N
 
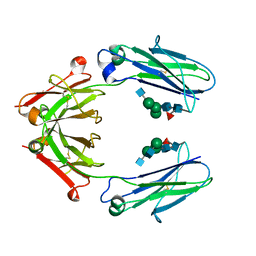 | | Crystal structure of a Fc fragment of Rhesus macaque (Macaca mulatta) IgG4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG4 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6NYH
 
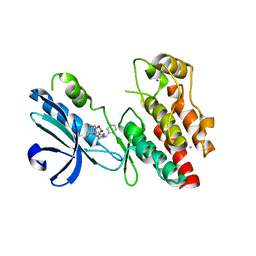 | | Structure of human RIPK1 kinase domain in complex with GNE684 | | Descriptor: | (5S)-N-[(3S)-7-methoxy-1-methyl-2-oxo-2,3,4,5-tetrahydro-1H-pyrido[3,4-b]azepin-3-yl]-5-phenyl-6,7-dihydro-5H-pyrrolo[1,2-b][1,2,4]triazole-2-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases.
Cell Death Differ., 27, 2020
|
|
6OKE
 
 | |
7M8J
 
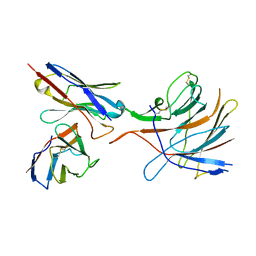 | | SARS-CoV-2 S-NTD + Fab CM25 | | Descriptor: | CM25 Fab - Heavy Chain, CM25 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes.
Science, 372, 2021
|
|
8FZB
 
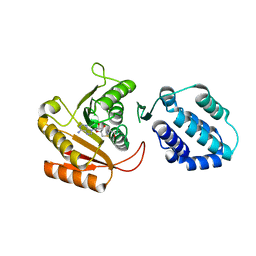 | | Crystal structure of human FAM86A | | Descriptor: | Protein-lysine N-methyltransferase EEF2KMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shao, Z, Lu, J, Song, J. | | Deposit date: | 2023-01-28 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The FAM86 domain of FAM86A confers substrate specificity to promote EEF2-Lys525 methylation.
J.Biol.Chem., 299, 2023
|
|
6P4L
 
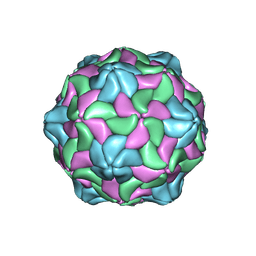 | |
6P4J
 
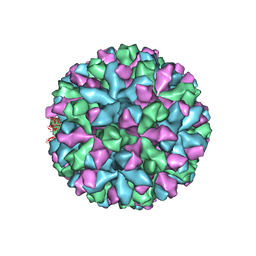 | | Mouse norovirus complexed with GCDCA | | Descriptor: | Capsid protein, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Smith, T.J, Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
6AVO
 
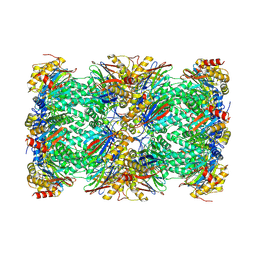 | | Cryo-EM structure of human immunoproteasome with a novel noncompetitive inhibitor that selectively inhibits activated lymphocytes | | Descriptor: | N~1~-{2-[([1,1'-biphenyl]-3-carbonyl)amino]ethyl}-N~4~-tert-butyl-N~2~-(3-phenylpropanoyl)-L-aspartamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Li, H, Santos, R, Bai, L. | | Deposit date: | 2017-09-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes.
Nat Commun, 8, 2017
|
|
6OKD
 
 | | Crystal Structure of human transferrin receptor in complex with a cystine-dense peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A TfR-Binding Cystine-Dense Peptide Promotes Blood-Brain Barrier Penetration of Bioactive Molecules.
J.Mol.Biol., 432, 2020
|
|
6ATK
 
 | |
8TXY
 
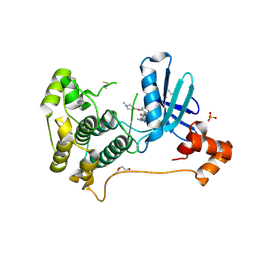 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6P4K
 
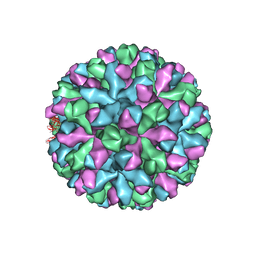 | | mouse norovirus complexed with TCA | | Descriptor: | Capsid protein, TAUROCHOLIC ACID | | Authors: | Smith, T.J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bile Salts Alter the Mouse Norovirus Capsid Conformation: Possible Implications for Cell Attachment and Immune Evasion.
J.Virol., 93, 2019
|
|
6MT3
 
 | |
6MTL
 
 | |
6MT5
 
 | |
