5DV4
 
 | |
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7TC3
 
 | | Human APE1 in the apo form | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7TC2
 
 | | Human APE1 in complex with 5-nitroindole-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
4QHD
 
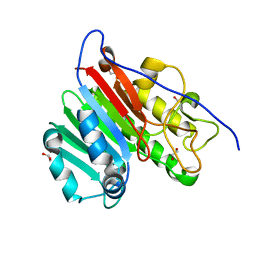 | | Crystal structure of apo human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | He, H, Chen, Q, Georgiadis, M.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
4QH9
 
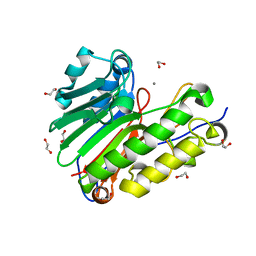 | | Crystal structure of Mn2+ bound human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase, MANGANESE (II) ION | | Authors: | Chen, Q, He, H, Georgiadis, M.M. | | Deposit date: | 2014-05-27 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
5DV2
 
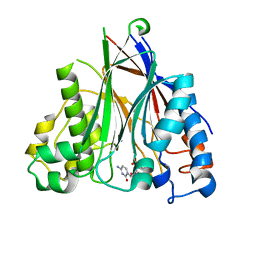 | |
7LPH
 
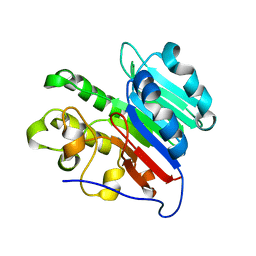 | | APE1 Mn-bound product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPJ
 
 | | APE1 Mn-bound phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPG
 
 | | APE1 product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPI
 
 | | APE1 phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
4QHE
 
 | | Crystal structure of Mg2+ bound human APE1 | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | He, H, Chen, Q, Georgiadis, M.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures reveal plasticity in the metal binding site of apurinic/apyrimidinic endonuclease I.
Biochemistry, 53, 2014
|
|
4RUW
 
 | |
1DE9
 
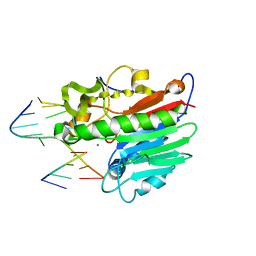 | | HUMAN APE1 ENDONUCLEASE WITH BOUND ABASIC DNA AND MN2+ ION | | Descriptor: | 5'-d(*CP*TP*AP*C)-3', 5'-d(*GP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-d(P*(3DR)P*GP*AP*TP*C)-3', ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
1DEW
 
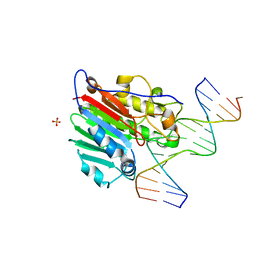 | | CRYSTAL STRUCTURE OF HUMAN APE1 BOUND TO ABASIC DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*(3DR)P*CP*GP*AP*CP*GP*AP*CP*G)-3', 5'-D(*GP*TP*CP*GP*TP*CP*GP*GP*GP*GP*AP*CP*GP*C)-3', MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE, ... | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination
Nature, 403, 2000
|
|
6NF0
 
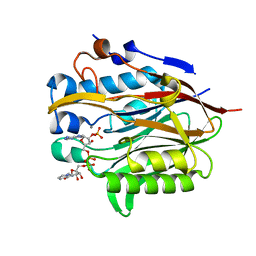 | | Nocturnin with bound NADPH substrate | | Descriptor: | CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nocturnin | | Authors: | Estrella, M.A, Du, J, Korennykh, A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The metabolites NADP+and NADPH are the targets of the circadian protein Nocturnin (Curled).
Nat Commun, 10, 2019
|
|
1DE8
 
 | | HUMAN APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1) BOUND TO ABASIC DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*CP*(3DR)P*GP*AP*TP*CP*G)-3'), MAJOR APURINIC/APYRIMIDINIC ENDONUCLEASE | | Authors: | Mol, C.D, Izumi, T, Mitra, S, Tainer, J.A. | | Deposit date: | 1999-11-13 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected
Nature, 403, 2000
|
|
1DNK
 
 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
5WN4
 
 | |
6MAL
 
 | |
5WN0
 
 | | APE1 exonuclease substrate complex with a C/G match | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
5WN2
 
 | |
6MKM
 
 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
5WN5
 
 | | APE1 exonuclease substrate complex with a C/T mismatch and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DV3))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
