8GRQ
 
 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GUI
 
 | | Bre1-nucleosome complex (Model I) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUJ
 
 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
5H7S
 
 | |
5J3X
 
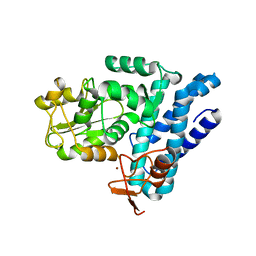 | | Structure of c-CBL Y371F | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, D.T, Buetow, L, Dou, H. | | Deposit date: | 2016-03-31 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation.
Bmc Biol., 14, 2016
|
|
5UDH
 
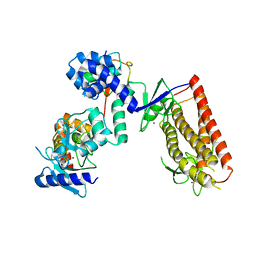 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5TTE
 
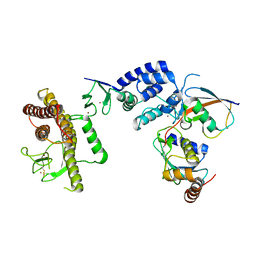 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
3VGO
 
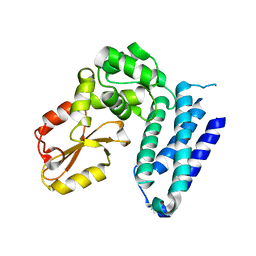 | |
6Y5N
 
 | | RING-DTC domain of Deltex1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase DTX1, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
6Y2X
 
 | | RING-DTC domains of Deltex 2, Form 2 | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-17 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
6Y3J
 
 | | RING-DTC domains of Deltex 2, bound to ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielssen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
6Y5P
 
 | | RING-DTC domain of Deltex1 bound to NAD | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase DTX1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into ADP-ribosylation of ubiquitin by Deltex family E3 ubiquitin ligases.
Sci Adv, 6, 2020
|
|
6Y22
 
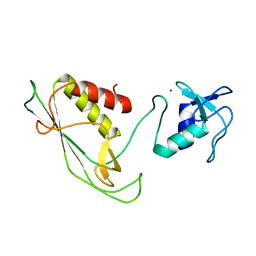 | | RING-DTC domains of Deltex 2, Form 1 | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Gabrielssen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | DELTEX2 C-terminal domain recognizes and recruits ADP-ribosylated proteins for ubiquitination.
Sci Adv, 6, 2020
|
|
7B5N
 
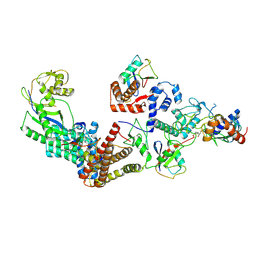 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-UBE2L3~Ub~ARIH1. | | Descriptor: | 5-azanylpentan-2-one, Cullin-1, E3 ubiquitin-protein ligase ARIH1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5S
 
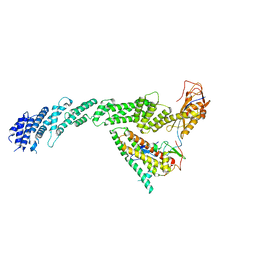 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-ARIH1 Ariadne. Transition State 1 | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase ARIH1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
7B5M
 
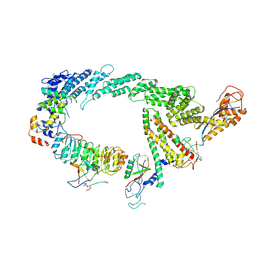 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
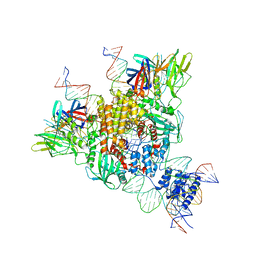 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEM
 
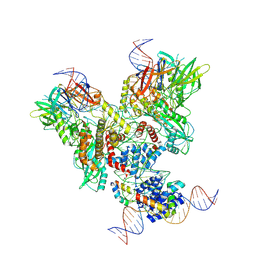 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3ZNI
 
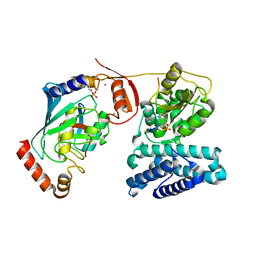 | | Structure of phosphoTyr363-Cbl-b - UbcH5B-Ub - ZAP-70 peptide complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE CBL-B, ... | | Authors: | Dou, H, Buetow, L, Sibbet, G.J, Cameron, K, Huang, D.T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Essentiality of a Non-Ring Element in Priming Donor Ubiquitin for Catalysis by a Monomeric E3.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6OES
 
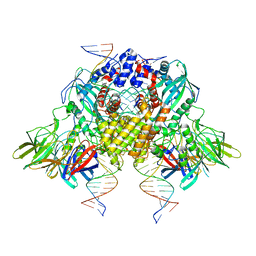 | | Cryo-EM structure of mouse RAG1/2 STC complex (without NBD domain) | | Descriptor: | CALCIUM ION, DNA (34-MER), DNA (35-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BBD
 
 | | Crystal structure of monoubiquitinated TRIM21 RING (Ub-RING) In complex with ubiquitin charged Ube2N (Ube2N~Ub) and Ube2V2 | | Descriptor: | Polyubiquitin-B,E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Kiss, L, Neuhaus, D, James, L.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination.
Nat Commun, 12, 2021
|
|
6OEP
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5VO0
 
 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | POTASSIUM ION, TNF receptor-associated factor 6, Ubiquitin, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
5VNZ
 
 | | Structure of a TRAF6-Ubc13~Ub complex | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2017-05-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Nat Commun, 8, 2017
|
|
