6R4X
 
 | |
6R55
 
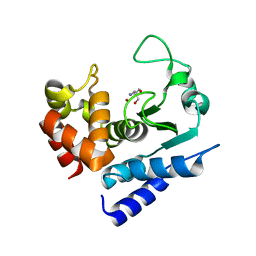 | | Crystal structure of PPEP-1(E184K) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
7NA8
 
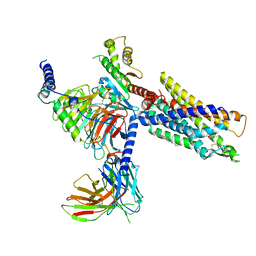 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | 1-(methanesulfonyl)-1'-(2-methyl-L-alanyl-O-benzyl-D-seryl)-1,2-dihydrospiro[indole-3,4'-piperidine], Antibody fragment, CHOLESTEROL, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
7NA7
 
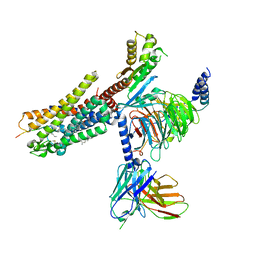 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | Antibody fragment, CHOLESTEROL, Ghrelin-27, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
5FCJ
 
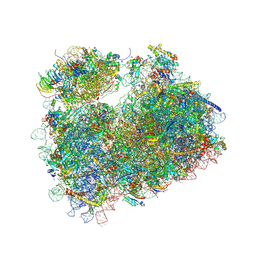 | | Structure of the anisomycin-containing uL3 W255C mutant 80S yeast ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Mailliot, J, Garreau de Loubresse, N, Yusupova, G, Dinman, J.D, Yusupov, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of the uL3 Mutant Ribosome: Illustration of the Importance of Ribosomal Proteins for Translation Efficiency.
J.Mol.Biol., 428, 2016
|
|
6R52
 
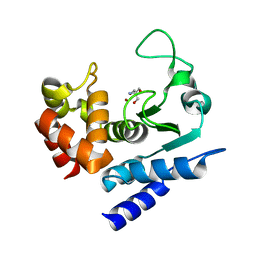 | | Crystal structure of PPEP-1(K101A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
6UJJ
 
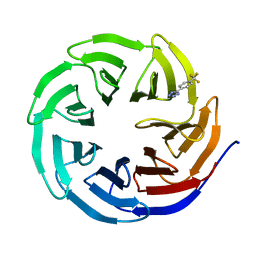 | |
6R92
 
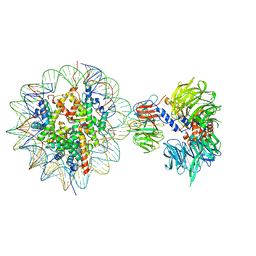 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R9Z
 
 | |
5FVM
 
 | | Cryo electron microscopy of a complex of Tor and Lst8 | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE TOR2, TARGET OF RAPAMYCIN COMPLEX SUBUNIT LST8 | | Authors: | Baretic, D, Berndt, A, Ohashi, Y, Johnson, C.M, Williams, R.L. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Tor Forms a Dimer Through an N-Terminal Helical Solenoid with a Complex Topology
Nat.Commun., 7, 2016
|
|
6UGM
 
 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
7MT2
 
 | | Mtb 70S initiation complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6UHZ
 
 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzotriazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.w. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
3J77
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
7MT3
 
 | | Mtb 70S with P/E tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7N40
 
 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
4ZH0
 
 | | Structure of Helicobacter pylori adhesin BabA determined by SeMet SAD | | Descriptor: | Outer membrane protein-adhesin | | Authors: | Howard, T.D, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
6O8X
 
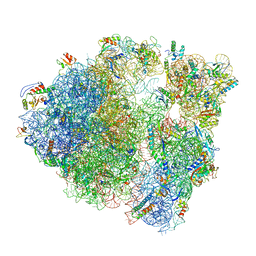 | |
7MT7
 
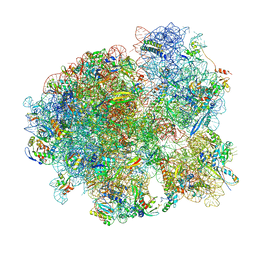 | | Mtb 70S with P and E site tRNAs | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6UHY
 
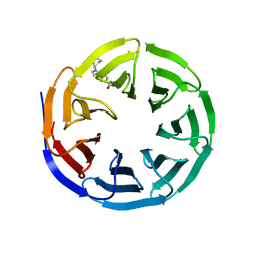 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzimidazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.W. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
3JCT
 
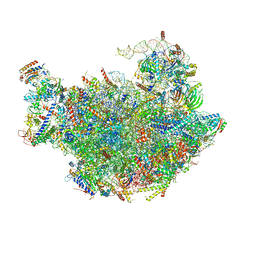 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
5IGO
 
 | |
6RBE
 
 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6R53
 
 | | Crystal structure of PPEP-1(K101R) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ZINC ION | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2019-03-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Molecular determinants of the mechanism and substrate specificity ofClostridium difficileproline-proline endopeptidase-1.
J.Biol.Chem., 294, 2019
|
|
3JZN
 
 | | Structure of EED in apo form | | Descriptor: | Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Ouyang, H, Qiu, W, MacKenzie, F, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
