8SES
 
 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
6MBK
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBL
 
 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, Second P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6XLK
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
3OUO
 
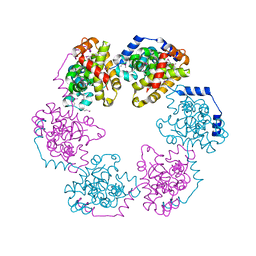 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-05-25 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
6Y4T
 
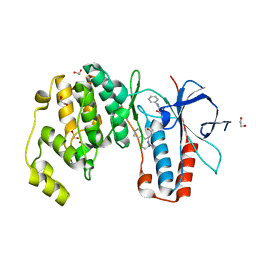 | | Crystal structure of p38 in complex with SR63. | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[[(2~{S})-butan-2-yl]amino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6YJC
 
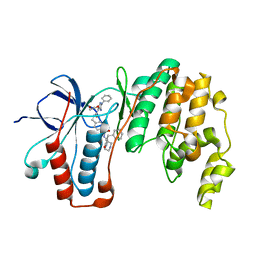 | | Crystal structure of p38alpha in complex with SR154 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-piperazin-1-yl-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74100935 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y4V
 
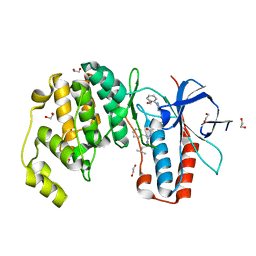 | | Crystal structure of p38 in complex with SR68 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6YK7
 
 | | Crystal structure of p38 in complex with SR43 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(ethylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
4ZF7
 
 | | Crystal structure of ferret interleukin-2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Interleukin 2, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Newman, J, McKinstry, W.J, Adams, T.E. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and functional characterisation of ferret interleukin-2.
Dev.Comp.Immunol., 55, 2015
|
|
6Y6V
 
 | | p38a bound with MCP-81 | | Descriptor: | 5-azanyl-~{N}-[[4-[[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-27 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y4W
 
 | | Crystal structure of p38 in complex with SR69 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(cyclohexylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
8PC5
 
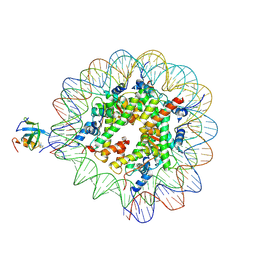 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PEO
 
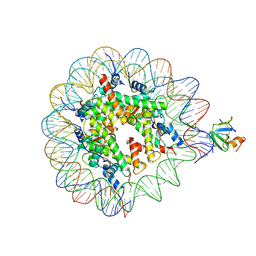 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8I53
 
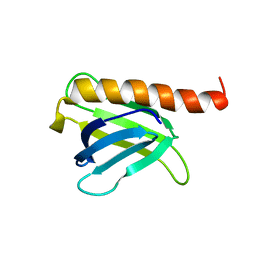 | |
8UIB
 
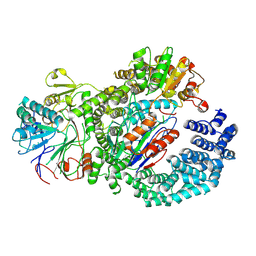 | | Structure of the human INTS9-INTS11-BRAT1 complex | | Descriptor: | BRCA1-associated ATM activator 1, Integrator complex subunit 11, Integrator complex subunit 9, ... | | Authors: | Lin, M, Tong, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cytoplasmic binding partners of the Integrator endonuclease INTS11 and its paralog CPSF73 are required for their nuclear function.
Mol.Cell, 84, 2024
|
|
6ZWP
 
 | | p38a bound with SR348 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[(4-fluorophenyl)amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Schroeder, M, Roehm, S, Knapp, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6Y4U
 
 | | Crystal structure of p38 in complex with SR65 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-(pentan-3-ylamino)butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7JTP
 
 | | Crystal structure of Protac MS67 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | Elongin-B, Elongin-C, GLYCEROL, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
6Y4X
 
 | | Crystal structure of p38 in complex with SR72 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-1-[2-(4-chlorophenyl)ethylamino]-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
7JTO
 
 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
