3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6G2I
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) at 5.9 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1, Breast cancer type 1 susceptibility protein | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
1J8E
 
 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
1IDC
 
 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
2QR2
 
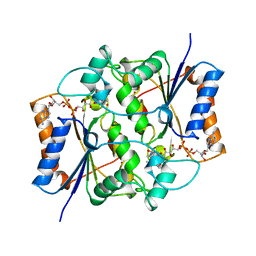 | | HUMAN QUINONE REDUCTASE TYPE 2, COMPLEX WITH MENADIONE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, PROTEIN (QUINONE REDUCTASE TYPE 2), ... | | Authors: | Foster, C, Bianchet, M.A, Talalay, P, Amzel, L.M. | | Deposit date: | 1999-04-19 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human quinone reductase type 2, a metalloflavoprotein.
Biochemistry, 38, 1999
|
|
3KMO
 
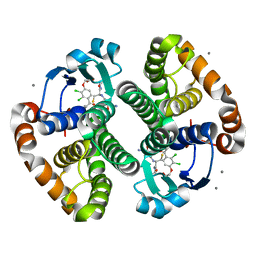 | |
4V91
 
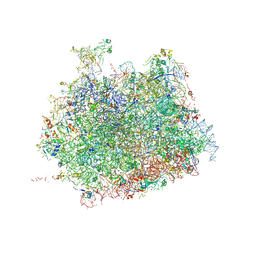 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
3KNS
 
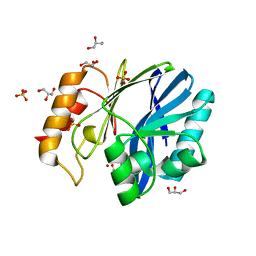 | | Bacillus cereus metallo-beta-lactamase Cys221Asp mutant, 20 mM Zn(II) | | Descriptor: | ACETIC ACID, Beta-lactamase 2, GLYCEROL, ... | | Authors: | Medrano Martin, F.J, Gonzalez, J.M, Vila, A.J. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Metallo-beta-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions.
Nat.Chem.Biol., 8, 2012
|
|
2QT4
 
 | | Atomic-resolution crystal structure of the natural form of Scytovirin | | Descriptor: | scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
2JGC
 
 | | Structure of the human eIF4E homologous protein, 4EHP without ligand bound | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 2, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E-BINDING PROTEIN 1 | | Authors: | Cameron, A.D, Rosettani, P, Knapp, S, Vismara, M.G, Rusconi, L. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms.
J. Mol. Biol., 368, 2007
|
|
3P5Q
 
 | | Ferric R-state human aquomethemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2010-10-10 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human R-state aquomethemoglobin at 2.0 A resolution
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6GD2
 
 | | Structure of HuR RRM3 in complex with RNA | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
1J4U
 
 | | Structure of Artocarpin Complexed with Me-alpha-Mannose | | Descriptor: | Artocarpin, methyl alpha-D-mannopyranoside | | Authors: | Pratap, J.V, Jeyaprakash, A.A, Rani, P.G, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2001-10-30 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of artocarpin, a Moraceae lectin with mannose specificity, and its complex with methyl-alpha-D-mannose: implications to the generation of carbohydrate specificity.
J.Mol.Biol., 317, 2002
|
|
1IDF
 
 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
2R33
 
 | |
4IZV
 
 | |
6FXU
 
 | |
2Z01
 
 | | Crystal structure of phosphoribosylaminoimidazole synthetase from Geobacillus kaustophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 2015
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
3KX4
 
 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant I401E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
3KYX
 
 | |
4GEL
 
 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
1J73
 
 | | Crystal structure of an unstable insulin analog with native activity. | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Wan, Z, Zhao, M, Nakagawa, S, Jia, W, Weiss, M.A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
4AB6
 
 | | Regulatory domain structure of NMB2055 (MetR), C103S C106S mutant, a LysR family regulator from N. meningitidis | | Descriptor: | SULFATE ION, TRANSCRIPTIONAL REGULATOR, LYSR FAMILY | | Authors: | Sainsbury, S, Ren, J, Saunders, N.J, Stuart, D.I, Owens, R.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Acta Crystallogr.,Sect.F, 68, 2012
|
|
