6GR3
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with AMPPNP | | Descriptor: | AMP PHOSPHORAMIDATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
4YAZ
 
 | | 3',3'-cGAMP riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4YB1
 
 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
6EA8
 
 | | Structure of VACV poxin in pre-reactive state with nonhydrolyzable 2'3' cGAMP | | Descriptor: | (2S,5R,7R,8R,10R,12aR,14R,15R,15aS,16R)-7-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-dihydroxy-2,10-disulfanyloctahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, O-[(1R,2R,3R)-5-{[(S)-{[(2R,3R,4R,5R)-2-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-4-hydroxy-5-(hydroxymethyl)tetrahydro furan-3-yl]oxy}(sulfanyl)phosphoryl]oxy}-1-(6-amino-9H-purin-9-yl)-1,2-dihydroxypentan-3-yl] dihydrogen (R)-phosphorothioate, Protein B2 | | Authors: | Eaglesham, J.B, Kranzusch, P.J. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling.
Nature, 566, 2019
|
|
2OOX
 
 | |
3QOP
 
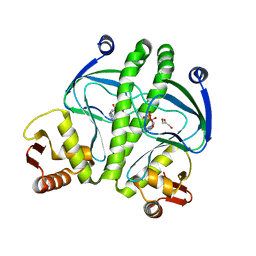 | |
6EP0
 
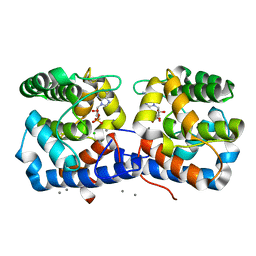 | |
2QRC
 
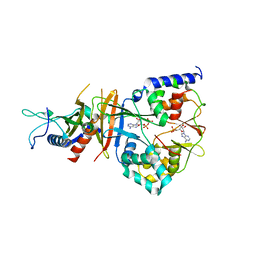 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
1LOI
 
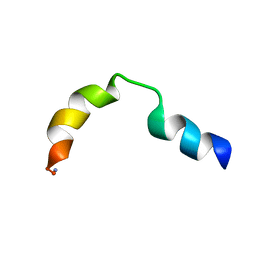 | | N-TERMINAL SPLICE REGION OF RAT C-AMP PHOSPHODIESTERASE, NMR, 7 STRUCTURES | | Descriptor: | CYCLIC 3',5'-AMP SPECIFIC PHOSPHODIESTERASE RD1 | | Authors: | Smith, K.J, Scotland, G, Beattie, J, Trayer, I.P, Houslay, M.D. | | Deposit date: | 1996-05-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the structure of the N-terminal splice region of the cyclic AMP-specific phosphodiesterase RD1 (RNPDE4A1) by 1H NMR and identification of the membrane association domain using chimeric constructs.
J.Biol.Chem., 271, 1996
|
|
5AHW
 
 | | Crystal structure of universal stress protein MSMEG_3811 in complex with cAMP | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Adolph, R.S, Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Universal Stress Protein (Usp) in Mycobacteria Binds Camp
J.Biol.Chem., 290, 2015
|
|
4FBP
 
 | | CONFORMATIONAL TRANSITION OF FRUCTOSE-1,6-BISPHOSPHATASE: STRUCTURE COMPARISON BETWEEN THE AMP COMPLEX (T FORM) AND THE FRUCTOSE 6-PHOSPHATE COMPLEX (R FORM) | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Ke, H, Zhang, Y, Liang, J.-Y, Lipscomb, W.N. | | Deposit date: | 1991-02-11 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational transition of fructose-1,6-bisphosphatase: structure comparison between the AMP complex (T form) and the fructose 6-phosphate complex (R form).
Biochemistry, 30, 1991
|
|
1RY2
 
 | |
5W10
 
 | | Lcd1 GAF domain in complex with cAMP ligand | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cGMP-specific phosphodiesterase | | Authors: | Guzzo, C.R, Maciel, N.K, Barbosa, A.S, Farah, C.S. | | Deposit date: | 2017-06-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Enzymatic Characterization of a cAMP-Dependent Diguanylate Cyclase from Pathogenic Leptospira Species.
J. Mol. Biol., 429, 2017
|
|
2RIF
 
 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
3U0Z
 
 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
6DT1
 
 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
5CFM
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' cGAMP, c[G(3', 5')pA(3', 5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
3U11
 
 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
4DG2
 
 | |
3U10
 
 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
4DG0
 
 | | Crystal structure of myristoylated WT catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP | | Descriptor: | MAGNESIUM ION, MYRISTIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Taylor, S.S. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase.
J.Mol.Biol., 422, 2012
|
|
4DFX
 
 | | Crystal structure of myristoylated K7C catalytic subunit of cAMP-dependent protein kinase in complex with SP20 and AMP-PNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Taylor, S.S. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase.
J.Mol.Biol., 422, 2012
|
|
4LEZ
 
 | | Structure of mouse cGAS bound to an 18bp DNA and cGAS product | | Descriptor: | 18bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION, ... | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
8BIK
 
 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
4K8V
 
 | | Structure of cyclic GMP-AMP Synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Gao, P, Wu, Y, Patel, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase.
Cell(Cambridge,Mass.), 153, 2013
|
|
