2CKY
 
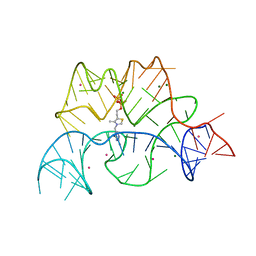 | | Structure of the Arabidopsis thaliana thiamine pyrophosphate riboswitch with its regulatory ligand | | Descriptor: | MAGNESIUM ION, NUCLEIC ACID, OSMIUM ION, ... | | Authors: | Thore, S, Leibundgut, M, Ban, N. | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Eukaryotic Thiamine Pyrophosphate Riboswitch with its Regulatory Ligand.
Science, 312, 2006
|
|
1O80
 
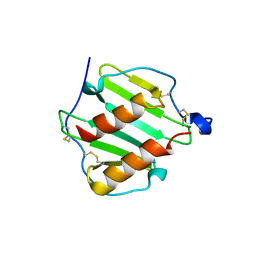 | |
1MDF
 
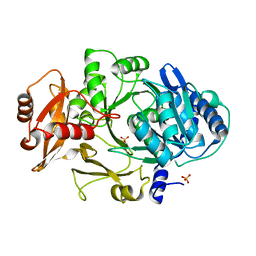 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3C0K
 
 | |
3DSJ
 
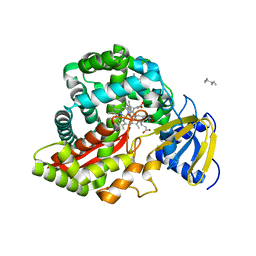 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase Variant (F137L) (At-AOS(F137L), cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOD at 1.60 A Resolution | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
3DS2
 
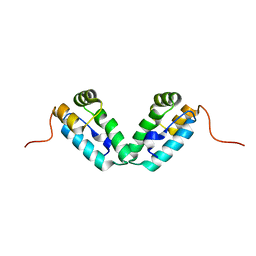 | | HIV-1 capsid C-terminal domain mutant (Y169A) | | Descriptor: | HIV-1 CAPSID PROTEIN | | Authors: | Vaney, M.-C, Igonet, S, Rey, F.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Residues in the HIV-1 Capsid Assembly Inhibitor Binding Site Are Essential for Maintaining the Assembly-competent Quaternary Structure of the Capsid Protein.
J.Biol.Chem., 283, 2008
|
|
4AIF
 
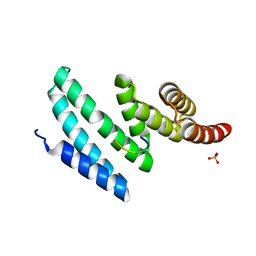 | | AIP TPR domain in complex with human Hsp90 peptide | | Descriptor: | AH RECEPTOR-INTERACTING PROTEIN, HEAT SHOCK PROTEIN HSP 90-ALPHA, SULFATE ION | | Authors: | Morgan, R.M.L, Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structure of the Tpr Domain of Aip: Lack of Client Protein Interaction with the C-Terminal Alpha-7 Helix of the Tpr Domain of Aip is Sufficient for Pituitary Adenoma Predisposition.
Plos One, 7, 2012
|
|
1M82
 
 | | SOLUTION STRUCTURE OF THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Descriptor: | RNA (25-MER): THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Authors: | Park, C.-J, Bae, S.-H, Lee, M.-K, Varani, G, Choi, B.-S. | | Deposit date: | 2002-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
NUCLEIC ACIDS RES., 31, 2003
|
|
1VK0
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
1MDB
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1O7Y
 
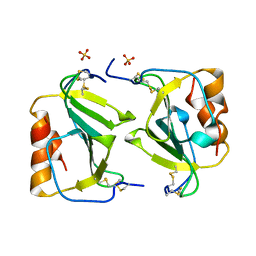 | | Crystal structure of IP-10 M-form | | Descriptor: | SMALL INDUCIBLE CYTOKINE B10, SULFATE ION | | Authors: | Swaminathan, G.J, Holloway, D.E, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-11-20 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Oligomeric Forms of the Ip-10/Cxcl10 Chemokine
Structure, 11, 2003
|
|
1GWH
 
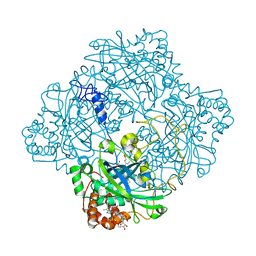 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase complexed with NADPH | | Descriptor: | ACETATE ION, Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2WFU
 
 | | Crystal structure of DILP5 variant DB | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
1GXT
 
 | | Hydrogenase Maturation Protein HypF "acylphosphatase-like" N-terminal domain (HypF-ACP) in complex with Sulfate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, HYDROGENASE MATURATION PROTEIN HYPF, ... | | Authors: | Rosano, C, Zuccotti, S, Stefani, M, Bucciantini, M, Ramponi, G, Bolognesi, M. | | Deposit date: | 2002-04-11 | | Release date: | 2002-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal Structure and Anion Binding in the Prokaryotic Hydrogenase Maturation Factor Hypf Acylphosphatase-Like Domain
J.Mol.Biol., 321, 2002
|
|
2W2D
 
 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3DSK
 
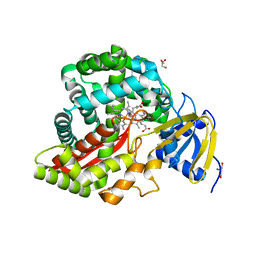 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase variant (F137L) (At-AOS(F137L), Cytochrome P450 74A, CYP74A) Complexed with 12R,13S-Vernolic Acid at 1.55 A Resolution | | Descriptor: | (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
2XSU
 
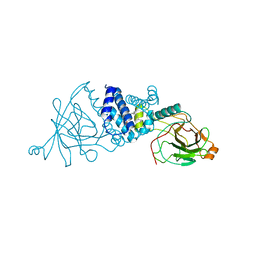 | | Crystal structure of the A72G mutant of Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
3BKD
 
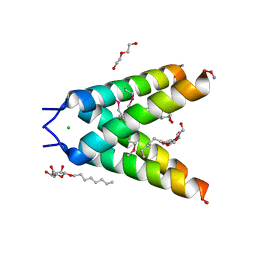 | | High resolution Crystal structure of Transmembrane domain of M2 protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Transmembrane Domain of Matrix protein M2, ... | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
1FZV
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PLACENTA GROWTH FACTOR-1 (PLGF-1), AN ANGIOGENIC PROTEIN AT 2.0A RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PLACENTA GROWTH FACTOR | | Authors: | Iyer, S, Leonidas, D.D, Swaminathan, G.J, Maglione, D, Battisti, M, Tucci, M, Persico, M.G, Acharya, K.R. | | Deposit date: | 2000-10-04 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human placenta growth factor-1 (PlGF-1), an angiogenic protein, at 2.0 A resolution.
J.Biol.Chem., 276, 2001
|
|
3DS1
 
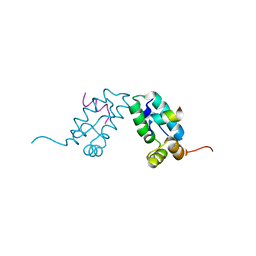 | |
2XIU
 
 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
3C7M
 
 | | Crystal structure of reduced DsbL | | Descriptor: | CADMIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Glockshuber, R, Grutter, M.G, Capitani, G. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli.
J.Mol.Biol., 380, 2008
|
|
2XSR
 
 | | Crystal structure of wild type Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2NQZ
 
 | | Trna-guanine transglycosylase (TGT) mutant in complex with 7-deaza-7-aminomethyl-guanine | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2006-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
