3IPU
 
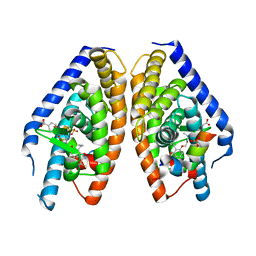 | | X-ray structure of benzisoxazole urea synthetic agonist bound to the LXR-alpha | | Descriptor: | 4-{[methyl(3-{[7-propyl-3-(trifluoromethyl)-1,2-benzisoxazol-6-yl]oxy}propyl)carbamoyl]amino}benzoic acid, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
5QPR
 
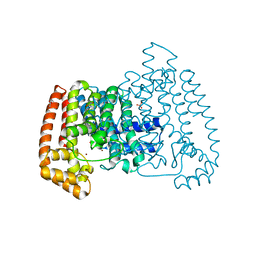 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with XST00001145b | | Descriptor: | 1-phenylmethoxyurea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
4ZA3
 
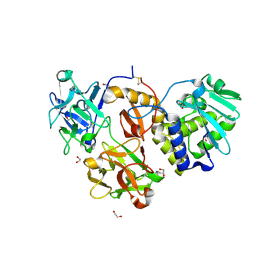 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd)-Native-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
7I4Q
 
 | | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre -- Crystal structure of Coxsackievirus A16 (G-10) 2A protease in complex with ASAP-0036805-001 (A71EV2A-x4735) | | Descriptor: | 1-[(2R)-2-(1H-imidazol-5-yl)pyrrolidin-1-yl]-2-[2-(methylsulfanyl)phenyl]ethan-1-one, DIMETHYL SULFOXIDE, Protease 2A, ... | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Godoy, A.S, Aschenbrenner, J.C, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Kenton, N, Tucker, J, DiPoto, M, Lee, A, Fearon, D, von Delft, F. | | Deposit date: | 2025-03-12 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Group deposition of Coxsackievirus A16 (G-10) 2A protease in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
1UMY
 
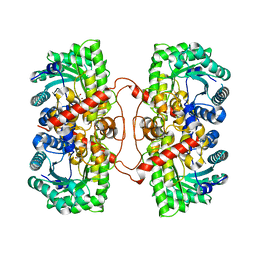 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
5QQ5
 
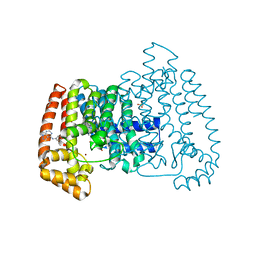 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with PKTTA024495b | | Descriptor: | 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5SBS
 
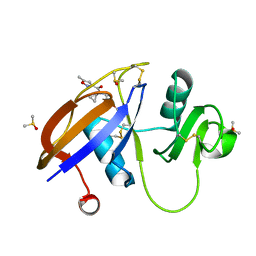 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z340495298 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.024 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
4CH8
 
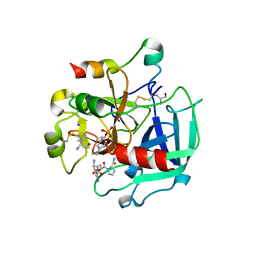 | | High-salt crystal structure of a thrombin-GpIbalpha peptide complex | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, PLATELET GLYCOPROTEIN IB ALPHA CHAIN, ... | | Authors: | Lechtenberg, B.C, Freund, S.M.V, Huntington, J.A. | | Deposit date: | 2013-11-29 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Gpibalpha Interacts Exclusively with Exosite II of Thrombin
J.Mol.Biol., 426, 2014
|
|
1UOQ
 
 | |
5SBW
 
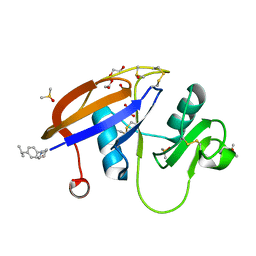 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z2856434874 | | Descriptor: | 1,2-ETHANEDIOL, 1-{[4-(propan-2-yl)phenyl]methyl}piperidin-4-ol, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
8CEH
 
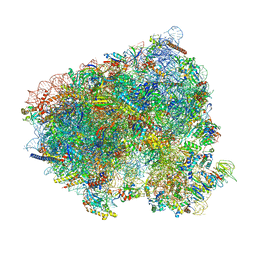 | | Translocation intermediate 4 (TI-4) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
5QPO
 
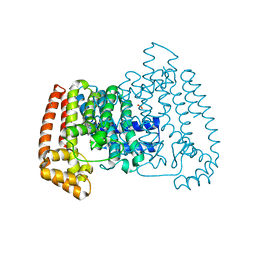 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000574a | | Descriptor: | 1-[(4-fluorophenyl)methyl]benzimidazole, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5SBZ
 
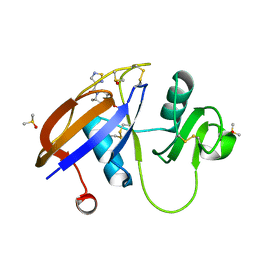 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z768399682 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC0
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z2856434899 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(thiophen-3-yl)methyl]piperidin-4-ol, CD44 antigen, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
2Y9R
 
 | |
5SC1
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z431807512 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
1NRR
 
 | |
2YDA
 
 | | Sulfolobus sulfataricus 2-keto-3-deoxygluconate aldolase Y103F,Y130F, A198F variant | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Crennell, S.J, Royer, S.F, Angelopoulou, M, Hough, D.W, Danson, M.J, Bull, S.D. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Sulfolobus Sulfataricus 2-Keto-3-Deoxygluconate Aldolase
To be Published
|
|
4CJL
 
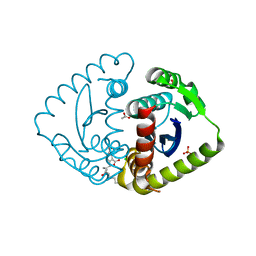 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[(2S)-2-{[(5-aminopentanoyl)amino]methyl}-4-methylpentyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-12-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CJV
 
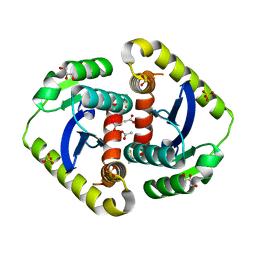 | |
8CB6
 
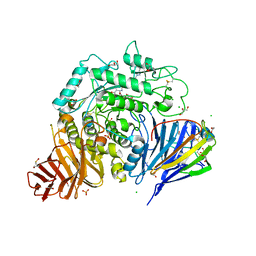 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in covalent complex with TAMRA tagged 1,6-Epi-cylcophellitol aziridine activity based probe | | Descriptor: | (1S,2R,3R,4R,5R)-5-[8-[4-(4-azanylbutyl)-1,2,3-triazol-1-yl]octylamino]-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
5SC5
 
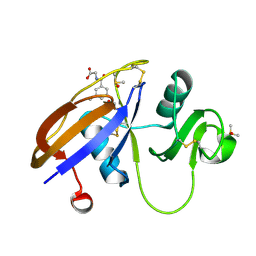 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with Z56827661 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
5SC6
 
 | | CD44 PanDDA analysis group deposition -- The hyaluronan-binding domain of CD44 in complex with POB0019 | | Descriptor: | 1,2-ETHANEDIOL, CD44 antigen, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bezerra, G.A, Koekemoer, L, von Delft, F, Bountra, C, Brennan, P.E, Gileadi, O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.327 Å) | | Cite: | CD44 PanDDA analysis group deposition
To Be Published
|
|
4R37
 
 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
