4V7H
 
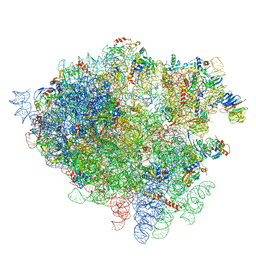 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
4VUB
 
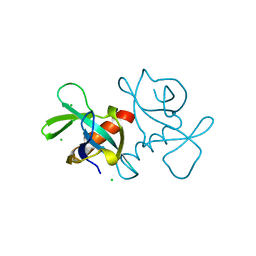 | | CCDB, A TOPOISOMERASE POISON FROM ESCHERICHIA COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
4V97
 
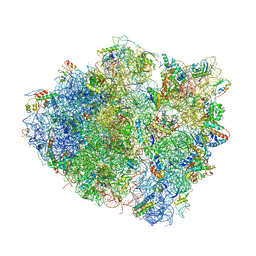 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2012-04-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V4N
 
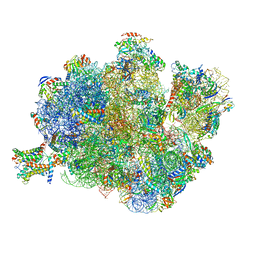 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
6M5Q
 
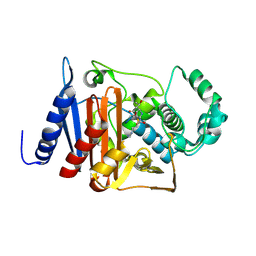 | | A class C beta-lactamase mutant - Y150F | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
7NDT
 
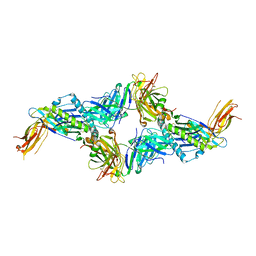 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDU
 
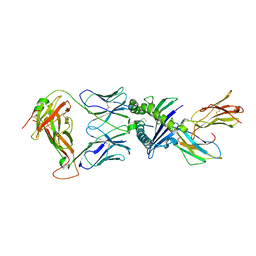 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDQ
 
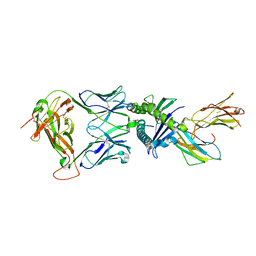 | | Gag:02 TCR in complex with HLA-E. | | Descriptor: | Beta-2-microglobulin, Gag6V, HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKZ
 
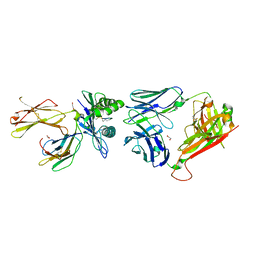 | | Crystal structure of InhA:01 TCR in complex with HLA-E (F116C) bound to InhA (53-61 H4C) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
8HFH
 
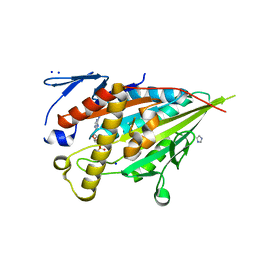 | | CENP-E motor domain in complex with AMPPNP and Mg2+ | | Descriptor: | Centromere-associated protein E, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Shibuya, A, Yokoyama, H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the motor domain of centromere-associated protein E in complex with a non-hydrolysable ATP analogue.
Febs Lett., 597, 2023
|
|
8G2B
 
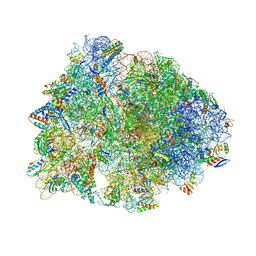 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAphe, and aminoacylated P-site fMet-tRNAmet at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G29
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2A
 
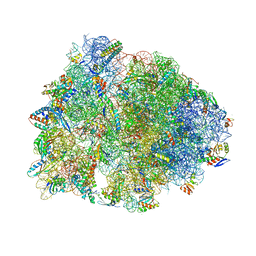 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, peptidyl P-site fMTHSMRC-tRNAmet, and deacylated E-site tRNAphe at 2.45A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
6ZKY
 
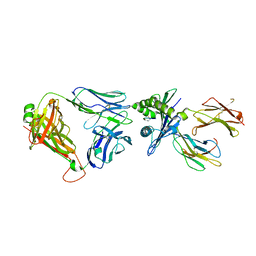 | | Crystal structure of InhA:01 TCR in complex with HLA-E (S147C) bound to InhA (53-61 H3C) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKW
 
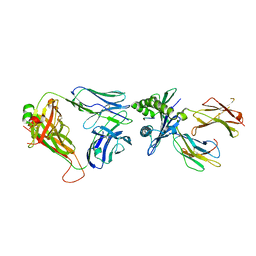 | | Crystal structure of InhA:01 TCR in complex with HLA-E bound to InhA (53-61) | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], HLA class I histocompatibility antigen, ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
6ZKX
 
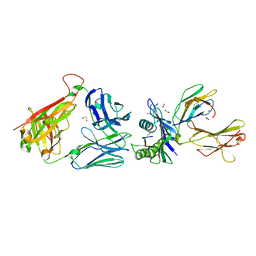 | | Crystal structure of InhA:01 TCR in complex with HLA-E (Y84C) bound to InhA (53-61 GCG) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Srikannathasan, V, Karuppiah, V, Robinson, R.A. | | Deposit date: | 2020-06-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
8G2C
 
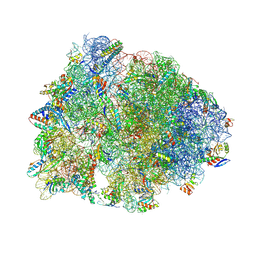 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2D
 
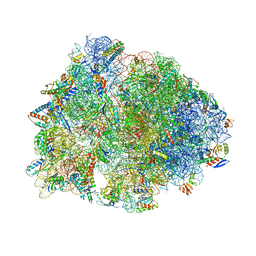 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
6NIP
 
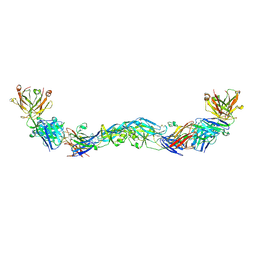 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ1 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, MZ1 Heavy chain, MZ1 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
6NIU
 
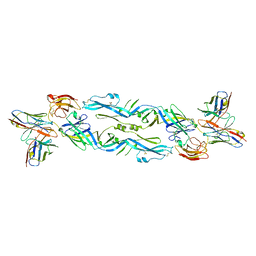 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ4 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, Human MZ4 Fab heavy chain, Human MZ4 Fab light chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.303 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat. Med., 26, 2020
|
|
6ZNA
 
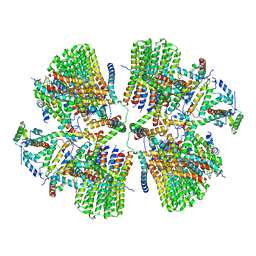 | | Porcine ATP synthase Fo domain | | Descriptor: | ATP synthase F(0) complex subunit C1, mitochondrial, ATP synthase g subunit, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZPO
 
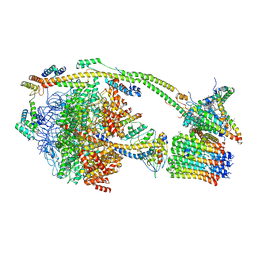 | | bovine ATP synthase monomer state 1 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8FV4
 
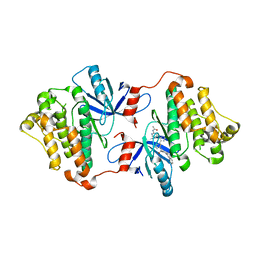 | | EGFR(T790M/V948R) in complex with compound 2 (LN5993) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(3P)-3-[(4P)-4-(2-acetamidopyridin-4-yl)-2-(methylsulfanyl)-1H-imidazol-5-yl]phenyl}-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepine-9-carboxamide, ... | | Authors: | Ogboo, B.C, Beyett, T.S, Eck, M.J, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
8FV3
 
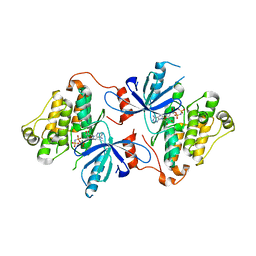 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
6ZQN
 
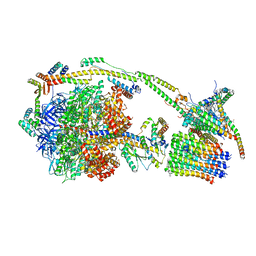 | | bovine ATP synthase monomer state 3 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
