2W1J
 
 | | CRYSTAL STRUCTURE OF SORTASE C-1 (SRTC-1) from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, PUTATIVE SORTASE | | Authors: | Manzano, C, Contreras-Martel, C, El Mortaji, L, Izore, T, Fenel, D, Vernet, T, Schoehn, G, Di Guilmi, A.M, Dessen, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Sortase-Mediated Pilus Fiber Biogenesis in Streptococcus Pneumoniae.
Structure, 16, 2008
|
|
7FKQ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04E09 from the F2X-Universal Library | | Descriptor: | 1-[(2-chloro-4-methoxyphenyl)methyl]-1H-1,2,4-triazole, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
7FO6
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H04 from the F2X-Universal Library | | Descriptor: | 1-[(2S)-2-methylmorpholin-4-yl]-2-(thiophen-3-yl)ethan-1-one, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
2C9Y
 
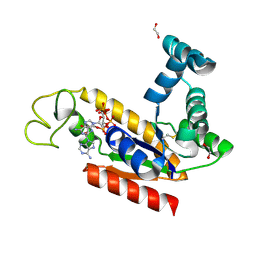 | | Structure of human adenylate kinase 2 | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE KINASE ISOENZYME 2, MITOCHONDRIAL, ... | | Authors: | Bunkoczi, G, Filippakopoulos, P, Debreczeni, J.E, Turnbull, A, Papagrigoriou, E, Savitsky, P, Colebrook, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human Adenylate Kinase 2
To be Published
|
|
5V7T
 
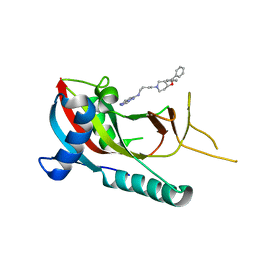 | | crystal structure of PARP14 bound to N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine inhibitor | | Descriptor: | N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Poly [ADP-ribose] polymerase 14 | | Authors: | saikatendu, k.s, Hirozane, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
6QFW
 
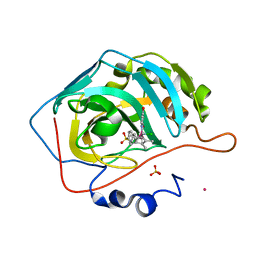 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 9) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[3-(8-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8$l^{4}-diaza-9$l^{7}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)propyl]benzenesulfonamide, Carbonic anhydrase 2, IRIDIUM ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
3ROC
 
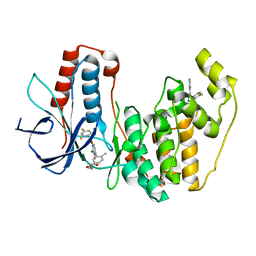 | | Crystal structure of human p38 alpha complexed with a pyrimidinone compound | | Descriptor: | 3-{5-chloro-4-[(2,4-difluorobenzyl)oxy]-6-oxopyrimidin-1(6H)-yl}-N-(2-hydroxyethyl)-4-methylbenzamide, 4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Xing, L. | | Deposit date: | 2011-04-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted N-aryl-6-pyrimidinones: A new class of potent, selective, and orally active p38 MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6M4W
 
 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9FQW
 
 | | Cryo-EM structure of MmCAT1 bound with FrMLV-RBD in the ornithine-bound inward-occluded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, High affinity cationic amino acid transporter 1,Green fluorescent protein, ... | | Authors: | Ye, M, Zhou, D, Pike, A.C.W, Wang, S, Wang, D, Bakshi, S, Brooke, L, Williams, E, Elkins, J, Stuart, D.I, Sauer, D.B. | | Deposit date: | 2024-06-17 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Amino acid and viral binding by the high-affinity Cationic Amino acid Transporter 1 (CAT1) from Mus musculus
To Be Published
|
|
7A0S
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
3U12
 
 | | The pleckstrin homology (PH) domain of USP37 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Dong, A, Nair, U.B, Wernimont, A, Walker, J.R, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The pleckstrin homology (PH) domain of USP37
To be Published
|
|
5ESJ
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
4P14
 
 | | Crystal structure of Canavalia brasiliensis (ConBr) complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, CALCIUM ION, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Silva-Filho, J.C, Teixeira, C.S, Cavada, B.S, Nascimento, K.S, Neto, I.L.B, Nobrega, R.B, Nagano, C.S, Sampaio, A.H, Holanda, E, Leal, R.B. | | Deposit date: | 2014-02-24 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Canavalia brasiliensis (ConBr) complexed with adenine
To Be Published
|
|
6M0K
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor 11b | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Liu, H, Rao, Z, Jiang, H. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease.
Science, 368, 2020
|
|
3TOS
 
 | | Crystal Structure of CalS11, Calicheamicin Methyltransferase | | Descriptor: | 1,2-ETHANEDIOL, CalS11, GLUTAMIC ACID, ... | | Authors: | Chang, A, Aceti, D.J, Beebe, E.T, Makino, S.-I, Wrobel, R.L, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2011-09-06 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of CalS11, Calicheamicin methyltransferase
To be Published
|
|
7K6R
 
 | |
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VZ3
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, T597C MUTANT | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Rea, D, Fulop, V. | | Deposit date: | 2004-05-14 | | Release date: | 2004-07-01 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Concerted Structural Changes in the Peptidase and the Propeller Domains of Prolyl Oligopeptidase are Required for Substrate Binding
J.Mol.Biol., 340, 2004
|
|
1VYK
 
 | | CRYSTAL STRUCTURE OF PSBQ PROTEIN OF PHOTOSYSTEM II FROM HIGHER PLANTS | | Descriptor: | ACETATE ION, OXYGEN-EVOLVING ENHANCER PROTEIN 3, ZINC ION | | Authors: | Hermoso, J.A, Balsera, M, De Las Rivas, J, Arellano, J.B. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.49 A Resolution Crystal Structure of Psbq from Photosystem II of Spinacia Oleracea Reveals a Ppii Structure in the N-Terminal Region.
J.Mol.Biol., 350, 2005
|
|
8CQ7
 
 | | Crystal structure of phyllanthoside bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-03 | | Release date: | 2024-09-11 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of read-through enhancement by aminoglycosides and mefloquine
Proc.Natl.Acad.Sci.USA, 2025
|
|
6M2J
 
 | | Uncommon structural features of rabbit MHC class I (RLA-A1) complexed with rabbit haemorrhagic disease virus (RHDV) derived peptide, VP60-1 | | Descriptor: | Beta-2-microglobulin, RLA class I histocompatibility antigen, alpha chain 19-1, ... | | Authors: | Zhang, Q.X, Liu, K.F, Yue, C, Zhang, D, Lu, D, Xiao, W.L, Liu, P.P, Zhao, Y.Z, Gao, G.L, Ding, C.M, Lyu, J.X, Liu, W.J. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strict Assembly Restriction of Peptides from Rabbit Hemorrhagic Disease Virus Presented by Rabbit Major Histocompatibility Complex Class I Molecule RLA-A1.
J.Virol., 94, 2020
|
|
