7EIK
 
 | | RD4-BD1 in complex with LT-872-297 | | Descriptor: | 1-[2-methyl-5-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-4-(1-methylpyrazol-4-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RD4-BD1 in complex with LT-872-297
To Be Published
|
|
5SGC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n3cn1c(nc(n1)N2CCOCC2)cc3NC(c4c(cnn4C)C(N5CCC5)=O)=O, micromolar IC50=0.004831 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-(morpholin-4-yl)[1,2,4]triazolo[1,5-c]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4B3L
 
 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2WDA
 
 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase in Complex with Chondroitin sulphate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
5SPV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with REAL250003774401 | | Descriptor: | 2-[methyl-[(9-oxidanylidene-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-3-yl)carbonyl]amino]ethanoic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1ZW9
 
 | |
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
7EJV
 
 | | The co-crystal structure of DYRK2 with YK-2-69 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [6-[[4-[2-(dimethylamino)-1,3-benzothiazol-6-yl]-5-fluoranyl-pyrimidin-2-yl]amino]pyridin-3-yl]-(4-ethylpiperazin-1-yl)methanone | | Authors: | Li, Z, Xiao, Y, Yuan, K, Kuang, W, Xiuquan, Y, Yang, P. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Nat Commun, 13, 2022
|
|
5SGK
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH [C@@H]2(Cc1nc(cn1CC2)c3ccccc3C)NC(=O)c5c(C(=O)N4CCC4)cnn5C, micromolar IC50=0.007851 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4R,7R)-2-(2-methylphenyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Bleicher, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5B6A
 
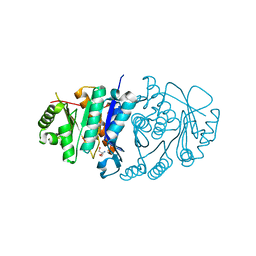 | | Structure of Pyridoxal Kinasefrom Pseudomonas Aeruginosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kim, M.I, Hong, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and catalytic mechanism of pyridoxal kinase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 478, 2016
|
|
6KSF
 
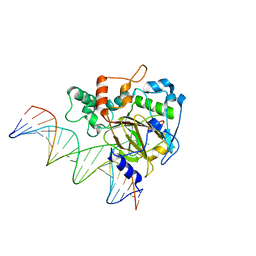 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | Authors: | Li, H, Zhang, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
3IE4
 
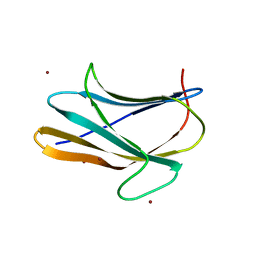 | | b-glucan binding domain of Drosophila GNBP3 defines a novel family of pattern recognition receptor | | Descriptor: | 1,2-ETHANEDIOL, Gram-Negative Binding Protein 3, ZINC ION | | Authors: | Mishima, Y, Coste, F, Kellenberger, C, Roussel, A. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors
To be Published
|
|
5SH2
 
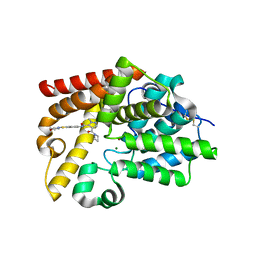 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C(c1c(cnn1C)C(N2CCC2)=O)(=O)Nc3cc4c(cc3)sc(n4)N5CCOCC5, micromolar IC50=0.001856 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(morpholin-4-yl)-1,3-benzothiazol-5-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4C2Y
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
5SFC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(cnn1C)C(Nc3nc2nc(cn2cc3)c4ccccc4)=O)C(N5CCC5)=O, micromolar IC50=2.191865 | | Descriptor: | 5-(azetidine-1-carbonyl)-1-methyl-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5FXV
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with N05859b | | Descriptor: | 1,2-ETHANEDIOL, 4-CARBOXYPIPERIDINE, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Srikannathasan, V, Oerum, S, Pearce, N.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with N05859B
To be Published
|
|
247L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5FXW
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with fumarate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Fumarate
To be Published
|
|
5SIP
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(n(nc1)C)C(=O)Nc3nc2nc(cn2cc3)c4ccccc4)C(N5CCC5)=O, micromolar IC50=0.0027158 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
251L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
5SKH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH C1=CN(N=C(C1=O)c2ccnn2c3cc(ccc3)F)c4cc(ccc4)OC(F)(F)F, micromolar IC50=0.043256 | | Descriptor: | 3-[1-(3-fluorophenyl)-1H-pyrazol-5-yl]-1-[3-(trifluoromethoxy)phenyl]pyridazin-4(1H)-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rogers-Evans, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
6KTM
 
 | |
5SKI
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(nc(c1ccccc1)nc2NCc4nn3c(cnc(c3n4)C)C)C, micromolar IC50=0.178156 | | Descriptor: | MAGNESIUM ION, N-{[(4R)-5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl]methyl}-1-methyl-3-phenyl-1H-1,2,4-triazol-5-amine, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SE8
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1(C)c(nc(n1)c2ccccc2)CCNC(c4c(C(N3CCC3)=O)cnn4C)=O, micromolar IC50=0.021263 | | Descriptor: | 4-(azetidine-1-carbonyl)-2-methyl-N-[2-(1-methyl-3-phenyl-1H-1,2,4-triazol-5-yl)ethyl]-2,5-dihydro-1H-pyrazole-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
