4CPX
 
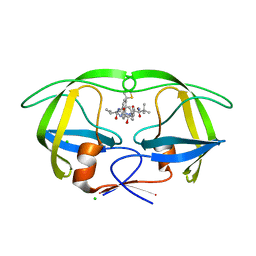 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[3-[(3~{R},6~{S},10~{Z})-3-oxidanyl-4,7-bis(oxidanylidene)-6-propan-2-yl-5,8-diazabicyclo[11.2.2]heptadeca-1(16),10,13(17),14-tetraen-3-yl]propyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of P1'-functionalized macrocyclic transition-state mimicking HIV-1 protease inhibitors encompassing a tertiary alcohol.
J. Med. Chem., 57, 2014
|
|
5M44
 
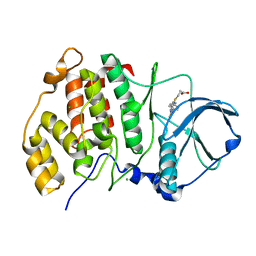 | | Complex structure of human protein kinase CK2 catalytic subunit with a thieno[2,3-d]pyrimidin inhibitor crystallized under high-salt conditions | | Descriptor: | 3-[5-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]sulfanylpropanoic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
4M2P
 
 | |
4CPR
 
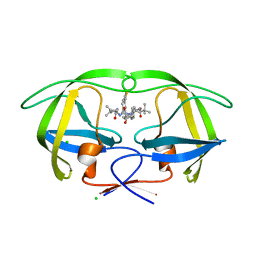 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[(4~{R})-5-[[(2~{S})-3-methyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-[(4-prop-2-enylphenyl)methyl]pentyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
2WBX
 
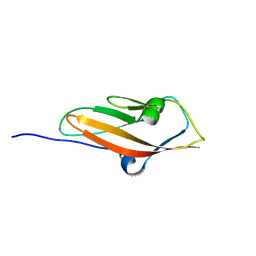 | | Crystal structure of mouse cadherin-23 EC1 | | Descriptor: | CADHERIN-23, CALCIUM ION | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-03-05 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
4CPU
 
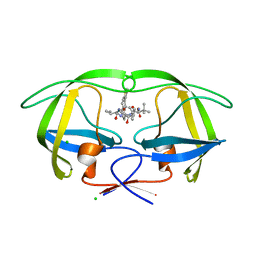 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4M4A
 
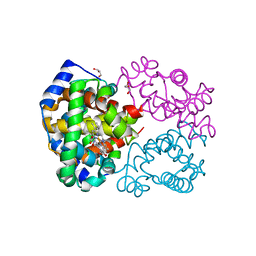 | | Human Hemoglobin Nitromethane Modified | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Yi, J, Guan, Y, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Degradation of human hemoglobin by organic C-nitroso compounds.
Chem.Commun.(Camb.), 49, 2013
|
|
4M4I
 
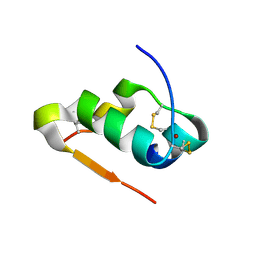 | | Radiation damage study of Cu T6-insulin - 0.12 MGy | | Descriptor: | COPPER (II) ION, Insulin | | Authors: | Frankaer, C.G, Harris, P, Stahl, K. | | Deposit date: | 2013-08-07 | | Release date: | 2014-01-15 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards accurate structural characterization of metal centres in protein crystals: the structures of Ni and Cu T6 bovine insulin derivatives.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LSD
 
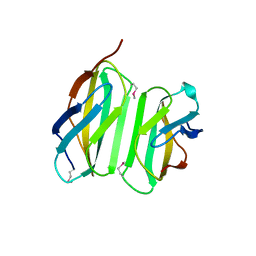 | | Myokine structure | | Descriptor: | Fibronectin type III domain-containing protein 5 | | Authors: | Schumacher, M.A, Ohashi, T, Shah, R.S, Chinnam, N, Erickson, H. | | Deposit date: | 2013-07-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation.
J.Biol.Chem., 288, 2013
|
|
4COE
 
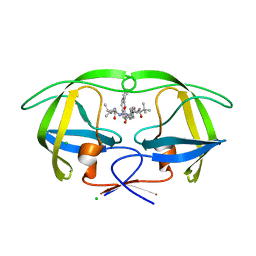 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl {(2S)-1-[2-(biphenyl-4-ylmethyl)-2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}hydrazinyl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
2VRP
 
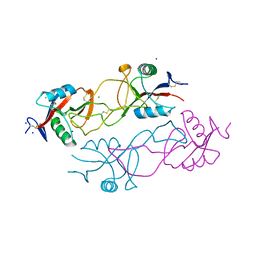 | | Structure of rhodocytin | | Descriptor: | AGGRETIN ALPHA CHAIN, AGGRETIN BETA CHAIN, CHLORIDE ION, ... | | Authors: | Watson, A.A, O'Callaghan, C.A. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of Rhodocytin, a Ligand for the Platelet-Activating Receptor Clec-2.
Protein Sci., 17, 2008
|
|
5LHS
 
 | | The ligand free catalytic domain of murine urokinase-type plasminogen activator | | Descriptor: | NICKEL (II) ION, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
4M6D
 
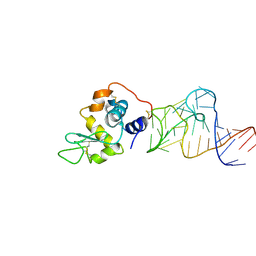 | | Crystal structure of the aptamer minF-lysozyme complex. | | Descriptor: | Lysozyme C, aptamer | | Authors: | Malashkevich, V.N, Padlan, F.C, Toro, R, Girvin, M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of the aptamer minF-lysozyme complex.
To be Published
|
|
2VZ7
 
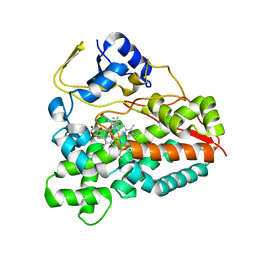 | | Crystal structure of the YC-17-bound PikC D50N mutant | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, S, Sherman, D.H, Podust, L.M. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Analysis of Transient and Catalytic Desosamine Binding Pockets in Cytochrome P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 284, 2009
|
|
8HYI
 
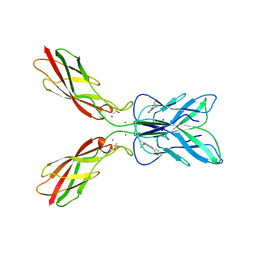 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine | | Descriptor: | 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Modulation of a conformational ensemble by a small molecule that inhibits key protein-protein interactions involved in cell adhesion.
Protein Sci., 32, 2023
|
|
2W5L
 
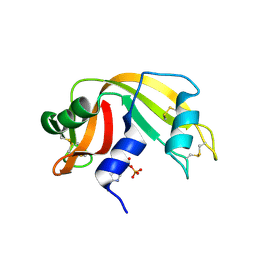 | | RNASE A-NADP COMPLEX | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
4LV6
 
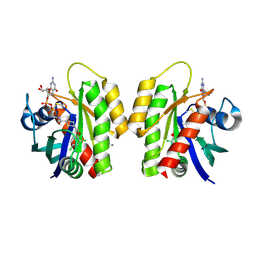 | | Crystal Structure of small molecule disulfide 4 covalently bound to K-Ras G12C | | Descriptor: | 1-[(2,4-dichlorophenoxy)acetyl]-N-(2-sulfanylethyl)piperidine-4-carboxamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-26 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
5LWE
 
 | | Crystal structure of the human CC chemokine receptor type 9 (CCR9) in complex with vercirnon | | Descriptor: | C-C chemokine receptor type 9, CHOLESTEROL, MALONATE ION, ... | | Authors: | Oswald, C, Rappas, M, Kean, J, Dore, A.S, Errey, J.C, Bennett, K, Deflorian, F, Christopher, J.A, Jazayeri, A, Mason, J.S, Congreve, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intracellular allosteric antagonism of the CCR9 receptor.
Nature, 540, 2016
|
|
4M8X
 
 | | GS-8374, a Novel Phosphonate-Containing Inhibitor of HIV-1 Protease, Effectively Inhibits HIV PR Mutants with Amino Acid Insertions | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Grantz saskova, K, Brynda, J, Rezacova, P, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-08-14 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GS-8374, a prototype phosphonate-containing inhibitor of HIV-1 protease, effectively inhibits protease mutants with amino acid insertions.
J.Virol., 88, 2014
|
|
5LWS
 
 | | Endothiapepsin in complex with fragment 177 and a derivative thereof | | Descriptor: | 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, 4-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazine, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LJA
 
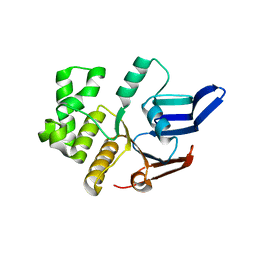 | | Structure of the E. coli MacB ABC domain (P6122) | | Descriptor: | Macrolide export ATP-binding/permease protein MacB | | Authors: | Crow, A. | | Deposit date: | 2016-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJE
 
 | | Crystal structure of holo human CRBP1/K40L,Q108L mutant | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
4M95
 
 | |
4M9Q
 
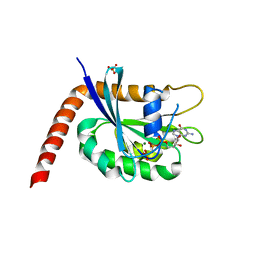 | | Crystal structure of C-terminally truncated Arl13B from Chlamydomonas rheinhardtii bound to GppNHp | | Descriptor: | ARF-like GTPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Miertzschke, M, Koerner, C, Spoerner, M, Wittinghofer, A. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the small G-protein Arl13B and implications for Joubert syndrome.
Biochem.J., 457, 2014
|
|
4LXJ
 
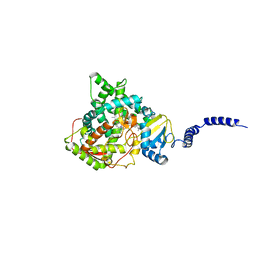 | | Saccharomyces cerevisiae lanosterol 14-alpha demethylase with lanosterol bound | | Descriptor: | LANOSTEROL, Lanosterol 14-alpha demethylase, OXYGEN MOLECULE, ... | | Authors: | Monk, B.C, Tomasiak, T.M, Keniya, M.V, Huschmann, F.U, Tyndall, J.D.A, O'Connell III, J.D, Cannon, R.D, McDonald, J, Rodriguez, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
