7RIM
 
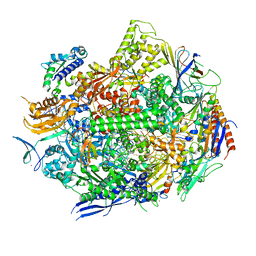 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4OQ2
 
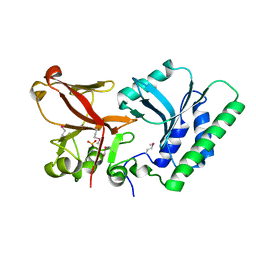 | | 5hmC specific restriction endonuclease PvuRTs1I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Restriction endonuclease PvuRts1 I | | Authors: | Kazrani, A.A, Kowalska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the 5hmC specific endonuclease PvuRts1I.
Nucleic Acids Res., 42, 2014
|
|
3I4X
 
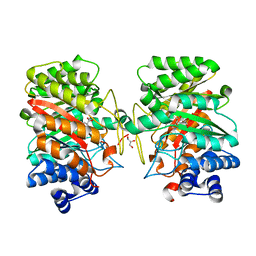 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus in complex with Trp and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3R4R
 
 | |
7RIP
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 1 soaked with CTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1L24
 
 | |
7RIY
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 soaked with UTP | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5XR8
 
 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
1L34
 
 | |
5MWK
 
 | | Glycoside hydrolase BT_0986 | | Descriptor: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 2, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
3EKS
 
 | | Crystal Structure of Monomeric Actin bound to Cytochalasin D | | Descriptor: | (3S,3aR,4S,6S,6aR,7E,10S,12R,13E,15R,15aR)-3-benzyl-6,12-dihydroxy-4,10,12-trimethyl-5-methylidene-1,11-dioxo-2,3,3a,4,5,6,6a,9,10,11,12,15-dodecahydro-1H-cycloundeca[d]isoindol-15-yl acetate, ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, ... | | Authors: | Nair, U.B, Joel, P.B, Wan, Q, Lowey, S, Rould, M.A, Trybus, K.M. | | Deposit date: | 2008-09-19 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of monomeric actin bound to cytochalasin D.
J.Mol.Biol., 384, 2008
|
|
6HCB
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.9 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Masternak, M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6QQV
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 2-[[6-(5-azanyl-1~{H}-pyrazol-3-yl)indol-1-yl]methyl]benzenecarbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6QR9
 
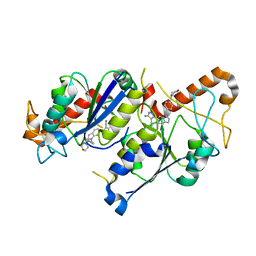 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(morpholin-4-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
3HY7
 
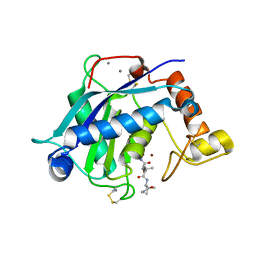 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
176L
 
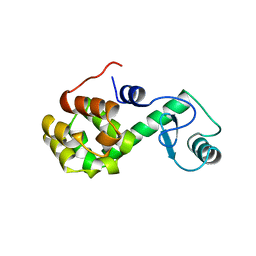 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L, Dubose, R, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
7UTZ
 
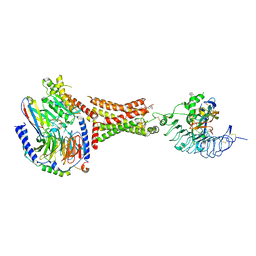 | | Human thyrotropin analog TR1402 bound to human Thyrotropin receptor in complex with miniGs399 (composite structure) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein hormones alpha chain analog TR1402, ... | | Authors: | Faust, B, Cheng, Y, Manglik, A. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Autoantibody mimicry of hormone action at the thyrotropin receptor.
Nature, 609, 2022
|
|
4ZKL
 
 | | Crystal structure of human histidine triad nucleotide-binding protein 1 (hHINT1) complexed with JB419 (AP4A analog) | | Descriptor: | (2R,3R,4S,5R)-2-(6-aminopurin-9-yl)-5-[2-[[(2S)-3-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-sulfanyl-phosphoryl]oxyethyl]oxolane-3,4-diol, ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Kaczmarek, R, Seda, A, Krakowiak, A, Baraniak, J, Nawrot, B. | | Deposit date: | 2015-04-30 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystallographic studies of the complex of human HINT1 protein with a non-hydrolyzable analog of Ap4A.
Int.J.Biol.Macromol., 87, 2016
|
|
6QR7
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(piperidin-1-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
6QZD
 
 | | HLA-DR1 with SGP Influenza Matrix Peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
6QMH
 
 | |
6HG2
 
 | | Hybrid dihydroorotase domain of human CAD with E. coli flexible loop, bound to fluoroorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Del Cano-Ochoa, F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization of the catalytic flexible loop in the dihydroorotase domain of the human multi-enzymatic protein CAD.
J. Biol. Chem., 293, 2018
|
|
4BWR
 
 | | Crystal structure of c5321: a protective antigen present in uropathogenic Escherichia coli strains displaying an SLR fold | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Urosev, D, Ferrer-Navarro, M, Pastorello, I, Cartocci, E, Costenaro, L, Zhulenkovs, D, Marechal, J.-D, Leonchiks, A, Reverter, D, Serino, L, Soriani, M, Daura, X. | | Deposit date: | 2013-07-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of C5321: A Protective Antigen Present in Uropathogenic Escherichia Coli Strains Displaying an Slr Fold.
Bmc Struct.Biol., 13, 2013
|
|
5H2P
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
9FZG
 
 | | Crystal structure of the hPXR-LBD in complex with compound JMV7035 | | Descriptor: | 2-(1-adamantyl)-~{N}-[2-[2-[2-oxidanylidene-2-[7-[1-(phenylmethyl)-5-[(2,4,6-trimethylphenyl)sulfonylamino]benzimidazol-2-yl]heptylamino]ethoxy]ethoxy]ethyl]ethanamide, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Delfosse, V, Bourguet, W. | | Deposit date: | 2024-07-05 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A two-in-one expression construct for biophysical and structural studies of the human pregnane X receptor ligand-binding domain, a pharmaceutical and environmental target.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
