7OSD
 
 | |
7OS8
 
 | | NMR SOLUTION STRUCTURE OF [Pro3,DLeu9]TL | | Descriptor: | PHE-VAL-PRO-TRP-PHE-SER-LYS-PHE-DLE-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment.
J.Med.Chem., 64, 2021
|
|
7YF7
 
 | |
1IYR
 
 | | NMR Structure Ensemble Of Dff-C Domain | | Descriptor: | DNA FRAGMENTATION FACTOR ALPHA SUBUNIT | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-09-05 | | Release date: | 2002-09-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Dff-C Domain of Dff45/Icad. A Structural Basis for the Regulation of Apoptotic DNA Fragmentation
J.Mol.Biol., 321, 2002
|
|
7PQW
 
 | | NMR solution structure of BCR4 | | Descriptor: | BCR4 | | Authors: | Loth, K, Paquet, F. | | Deposit date: | 2021-09-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Aphid BCR4 Structure and Activity Uncover a New Defensin Peptide Superfamily.
Int J Mol Sci, 23, 2022
|
|
8B6X
 
 | |
8B6Y
 
 | |
2Z2G
 
 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
7BEV
 
 | |
6NZ2
 
 | |
7D12
 
 | | NMR solution structures of CAG RNA-DB213 binding complex | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*CP*UP*UP*CP*GP*GP*CP*AP*GP*CP*AP*GP*C)-3'), SODIUM ION | | Authors: | Chan, H.Y.E, Guo, P. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XTH
 
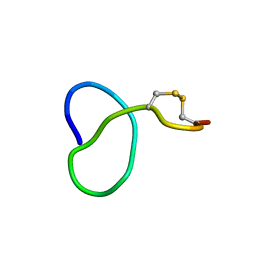 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A1 | | Authors: | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTI
 
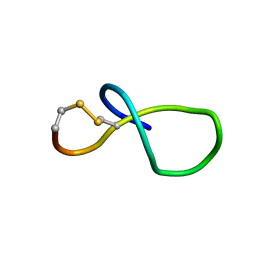 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A2 | | Authors: | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
7E4E
 
 | |
2Z2H
 
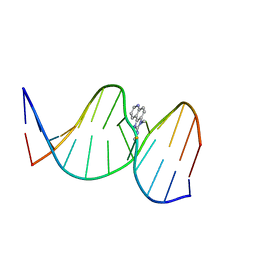 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | Authors: | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2BJC
 
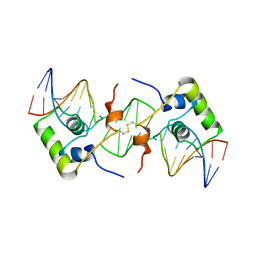 | | NMR structure of a protein-DNA complex of an altered specificity mutant of the lac repressor headpiece that mimics the gal repressor | | Descriptor: | 5'-D(*GP*AP*AP*TP*TP*GP*TP*AP*AP*GP *CP*GP*CP*TP*TP*AP*CP*AP*AP*TP*TP*C)-3', LACTOSE OPERON REPRESSOR | | Authors: | Salinas, R.K, Folkers, G.E, Bonvin, A.M.J.J, Das, D, Boelens, R, Kaptein, R. | | Deposit date: | 2005-02-01 | | Release date: | 2005-10-18 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Altered Specificity in DNA Binding by the Lac Repressor: A Mutant Lac Headpiece that Mimics the Gal Repressor
Chembiochem, 6, 2005
|
|
7NHZ
 
 | |
7A0O
 
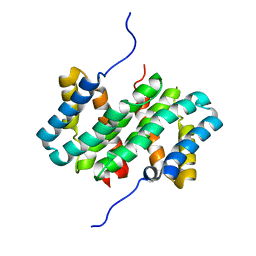 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 5.5 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7A0I
 
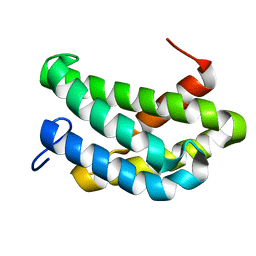 | | NMR structure of flagelliform spidroin (FlagSp) N-terminal domain from Trichonephila clavipes at pH 7.2 | | Descriptor: | Flagelliform spidroin variant 1 | | Authors: | Sarr, M, Kitoka, K, Walsh-White, K.-A, Kaldmae, M, Landreh, M, Rising, A, Johansson, J, Jaudzems, K, Kronqvist, N. | | Deposit date: | 2020-08-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The dimerization mechanism of the N-terminal domain of spider silk proteins is conserved despite extensive sequence divergence.
J.Biol.Chem., 298, 2022
|
|
7N1Z
 
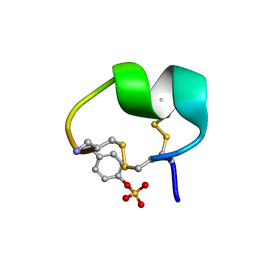 | | NMR structure of native PnIA | | Descriptor: | Alpha-conotoxin PnIA | | Authors: | Conibear, A.C, Rosengren, K.J, Lee, H.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Posttranslational modifications of alpha-conotoxins: sulfotyrosine and C-terminal amidation stabilise structures and increase acetylcholine receptor binding.
Rsc Med Chem, 12, 2021
|
|
7L2G
 
 | |
5T4R
 
 | |
6SAI
 
 | |
6M6K
 
 | |
6LNZ
 
 | |
