5MU6
 
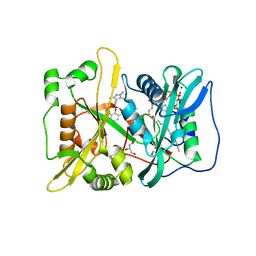 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and IMP-1088 inhibitor bound | | Descriptor: | 1-[5-[3,4-bis(fluoranyl)-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Perez-Dorado, I, Bell, A.S, Tate, E.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus.
Nat Chem, 10, 2018
|
|
9EMO
 
 | |
3VB7
 
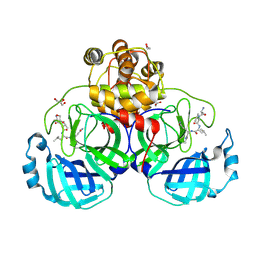 | | Crystal structure of SARS-CoV 3C-like protease with M4Z | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, GLYCEROL, ... | | Authors: | Chuck, C.P, Wong, K.B. | | Deposit date: | 2011-12-31 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases
Eur.J.Med.Chem., 59C, 2012
|
|
6P0E
 
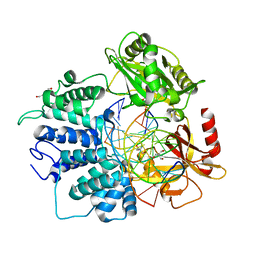 | | Human DNA Ligase 1 (E346A,E592A) bound to adenylated DNA containing an 8-oxo guanine:adenine base-pair | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
3HFB
 
 | |
7VG2
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VG3
 
 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a RNA forward strand (5'-phosphorylated), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
5GVM
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS557 | | Descriptor: | 2-[3-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]phenyl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G. | | Deposit date: | 2016-09-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Antimalarial Inhibitors Targeting Serine Hydroxymethyltransferase (SHMT) with in Vivo Efficacy and Analysis of their Binding Mode Based on X-ray Cocrystal Structures
J. Med. Chem., 60, 2017
|
|
6S70
 
 | | Crystal structure of CARM1 in complex with inhibitor UM251 | | Descriptor: | 1-[5-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]pentyl]guanidine, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S2B
 
 | | Structure of beta-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosyl-erythritol | | Descriptor: | (2~{S},3~{R})-4-[(2~{R},3~{S},4~{S},5~{R})-2,5-bis(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxybutane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New insights into the molecular mechanism behind mannitol and erythritol fructosylation by beta-fructofuranosidase from Schwanniomyces occidentalis.
Sci Rep, 11, 2021
|
|
3HIV
 
 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8B9Z
 
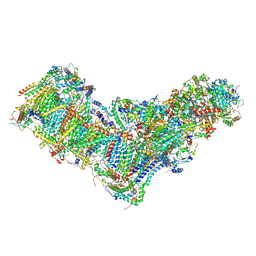 | | Drosophila melanogaster complex I in the Active state (Dm1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Agip, A.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
7DQQ
 
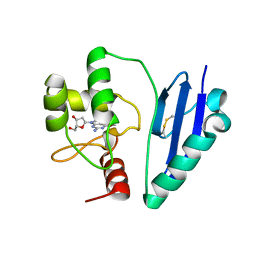 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant in complex with O6-methyldeoxyguanosine | | Descriptor: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8GHE
 
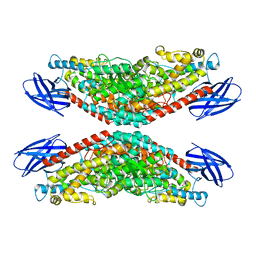 | | The structure of h12-LOX in tetrameric form bound to endogenous inhibitor oleoyl-CoA | | Descriptor: | FE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX12, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Black, K.A, Mobbs, J.I, Venugopal, H, Thal, D.M, Glukhova, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Cryo-EM structures of human arachidonate 12S-lipoxygenase bound to endogenous and exogenous inhibitors.
Blood, 142, 2023
|
|
6PB5
 
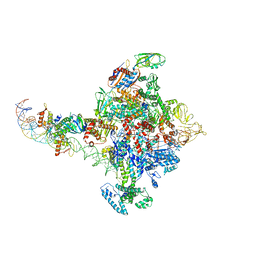 | |
4Z1O
 
 | |
3Q8E
 
 | | Crystal structure of Protective Antigen W346F (pH 8.5) | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
8YRK
 
 | | Tubulin-Compound KY216: stathmin-like domain complex | | Descriptor: | (11R,16S)-11-(3,5-dichloro-4-methoxyphenyl)-4,7-dioxa-12,14-diazatetracyclo[8.7.0.03,8.012,16]heptadeca-1,3(8),9-triene-13,15-dione, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Xia, Y.Z, He, X.Y. | | Deposit date: | 2024-03-21 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Tubulin-Compound KY216: stathmin-like domain complex
To Be Published
|
|
3VF5
 
 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
8W07
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-402 | | Descriptor: | (1R,2S,4R)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-cyclohexyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.K, Nettles, K.W. | | Deposit date: | 2024-02-13 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1WKW
 
 | | Crystal structure of the ternary complex of eIF4E-m7GpppA-4EBP1 peptide | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E binding protein 1, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | Authors: | Tomoo, K, Matsushita, Y, Fujisaki, H, Shen, X, Miyagawa, H, Kitamura, K, Miura, K, Ishida, T. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods
Biochim.Biophys.Acta, 1753, 2005
|
|
6SB2
 
 | | cryo-EM structure of mTORC1 bound to active RagA/C GTPases | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Ras-related GTP-binding protein A, ... | | Authors: | Anandapadamanaban, M, Berndt, A, Masson, G.R, Perisic, O, Williams, R.L. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science, 366, 2019
|
|
5QI6
 
 | | PanDDA analysis group deposition -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000597a | | Descriptor: | 4-[(5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)amino]phenol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3QUN
 
 | | Crystal structure of fosfomycin resistance kinase FomA from Streptomyces wedmorensis complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FomA protein, GLYCEROL, ... | | Authors: | Pakhomova, S, Bartlett, S.G, Doerner, P.A, Newcomer, M.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical insights into the mechanism of fosfomycin phosphorylation by fosfomycin resistance kinase FomA.
Biochemistry, 50, 2011
|
|
