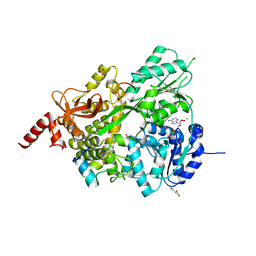
8EK4
 
 | |
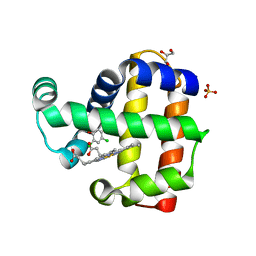
8EK3
 
 | |
8EK1
 
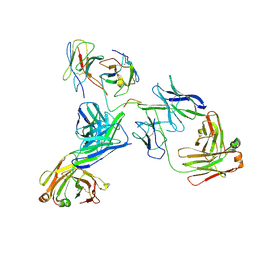 | |
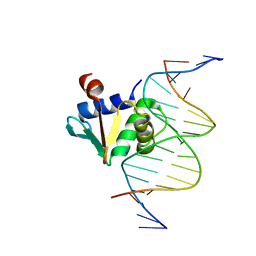
8EJN
 
 | | Structure of dehaloperoxidase A in complex with 2,4-dichlorophenol | | Descriptor: | 2,4-dichlorophenol, DIMETHYL SULFOXIDE, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Franzen, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Comparative study of the binding and activation of 2,4-dichlorophenol by dehaloperoxidase A and B.
J.Inorg.Biochem., 247, 2023
|
|
8EJM
 
 | |
8EJJ
 
 | | Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJH
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab | | Descriptor: | 12.1F Fab heavy chain, 12.1F Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
8EJF
 
 | | Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJE
 
 | | Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein GP1, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJ8
 
 | |
8EJ7
 
 | |
8EJ6
 
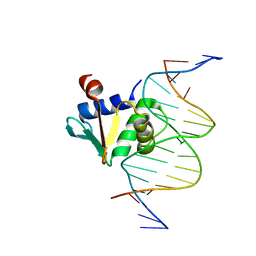 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EJ4
 
 | | Cryo-EM structure of the active NLRP3 inflammasome disk | | Descriptor: | MAGNESIUM ION, NACHT, LRR and PYD domains-containing protein 3, ... | | Authors: | Hao, W, Le, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of the active NLRP3 inflammasome disc.
Nature, 613, 2023
|
|
8EJ0
 
 | |
8EIZ
 
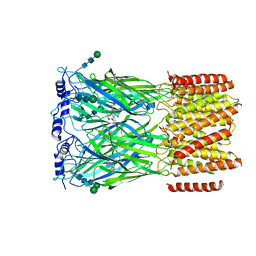 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
8EIS
 
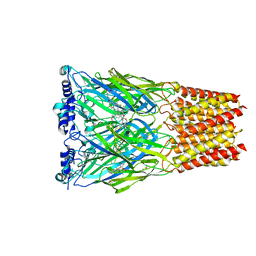 | | Cryo-EM structure of octopus sensory receptor CRT1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Octopus sensory receptor | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
8EIP
 
 | |
8EIO
 
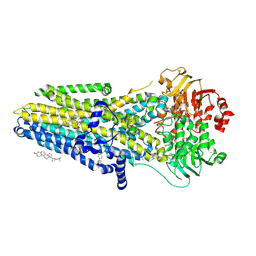 | | The complex of phosphorylated human delta F508 cystic fibrosis transmembrane conductance regulator (CFTR) with elexacaftor (VX-445), lumacaftor (VX-809) and ATP/Mg | | Descriptor: | (6P)-N-(1,3-dimethyl-1H-pyrazole-4-sulfonyl)-6-[3-(3,3,3-trifluoro-2,2-dimethylpropoxy)-1H-pyrazol-1-yl]-2-[(4S)-2,2,4-trimethylpyrrolidin-1-yl]pyridine-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, ... | | Authors: | Fiedorczuk, K, Chen, J. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular structures reveal synergistic rescue of Delta 508 CFTR by Trikafta modulators.
Science, 378, 2022
|
|
8EIN
 
 | |
8EIM
 
 | |
8EIL
 
 | | C-Terminal Domain of BrxL from Acinetobacter BREX type I phage restriction system | | Descriptor: | MALONIC ACID, Protease Lon-related BREX system protein BrxL, SUCCINIC ACID | | Authors: | Doyle, L.A, Stoddard, B.L, Kaiser, B. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure, substrate binding and activity of a unique AAA+ protein: the BrxL phage restriction factor.
Nucleic Acids Res., 51, 2023
|
|
8EIF
 
 | |
