8ZVV
 
 | |
8ZXG
 
 | | Crystal structure of Paraoxonase from Bacillus sp. strain S3wahi | | Descriptor: | GLYCEROL, MAGNESIUM ION, Paraoxonase from Bacillus sp. strain S3wahi, ... | | Authors: | Azman, A.A, Muhd Noor, N.D, Leow, A.T.C, Mohd Noor, S.A, Mohamad Ali, M.S. | | Deposit date: | 2024-06-14 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Crystal structure analysis of Paraoxonase from Bacillus sp. strain S3wahi
To Be Published
|
|
2HZK
 
 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its open form | | Descriptor: | GLYCEROL, TRAP-T family sorbitol/mannitol transporter, periplasmic binding protein, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
8ZXB
 
 | | Structure of aminotransferase from Mycolicibacterium neoaurum in complex with PLP | | Descriptor: | Branched-chain amino acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wei, H, Cong, L, Wang, J, You, S, Liu, W. | | Deposit date: | 2024-06-14 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of aminotransferase from Mycolicibacterium neoaurum in complex with PLP
To Be Published
|
|
5JRS
 
 | |
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
8ZXY
 
 | | sweet protein MNEI-Mut 6-2 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-15 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
8ZU4
 
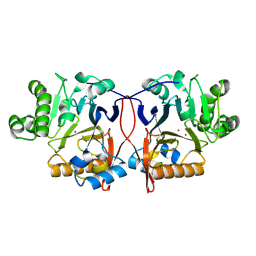 | |
2I4G
 
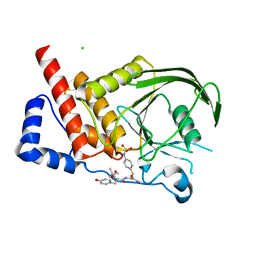 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with a sulfamic acid (soaking experiment) | | Descriptor: | CHLORIDE ION, N-(TERT-BUTOXYCARBONYL)-L-TYROSYL-N-METHYL-4-(SULFOAMINO)-L-PHENYLALANINAMIDE, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8ZXJ
 
 | | Sweet protein MNEI-Mut 6-1 | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | You, T.J, Liu, S. | | Deposit date: | 2024-06-14 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Exceptional Thermal Stability of the Boiling-Resistant Sweet Protein MNEI.
J.Agric.Food Chem., 73, 2025
|
|
8ZXI
 
 | |
5JW5
 
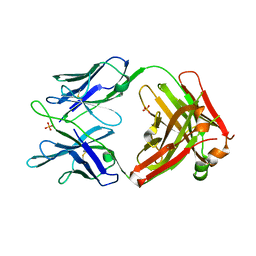 | | Structure of MEDI8852 Fab Fragment | | Descriptor: | MEDI8852 Heavy chain, MEDI8852 Light chain, PHOSPHATE ION | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
2I0G
 
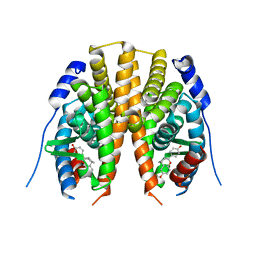 | | Benzopyrans are Selective Estrogen Receptor beta Agonists (SERBAs) with Novel Activity in Models of Benign Prostatic Hyperplasia | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Benzopyrans are selective estrogen receptor Beta agonists with novel activity in models of benign prostatic hyperplasia.
J.Med.Chem., 49, 2006
|
|
8HO8
 
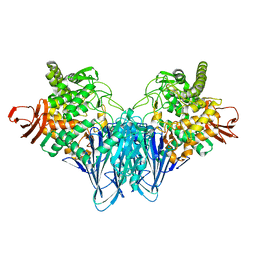 | |
7RTK
 
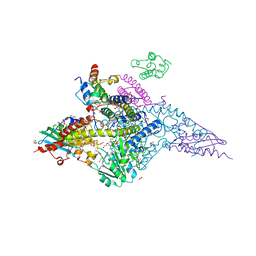 | | Structure of the (NIAU)2 complex with N-terminal mutation of ISCU2 Y35D at 2.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2021-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
2I0T
 
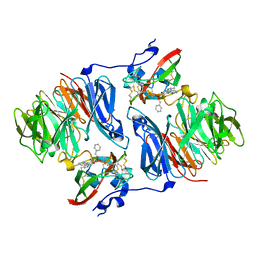 | |
8ZW1
 
 | |
6LFL
 
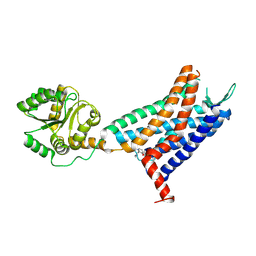 | | Crystal structure of a class A GPCR | | Descriptor: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, C-X-C chemokine receptor type 2,GlgA glycogen synthase,C-X-C chemokine receptor type 2 | | Authors: | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
8ZWQ
 
 | | pseudorabies virus dUTPase structure | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, MAGNESIUM ION, dUTPase | | Authors: | Wang, Y. | | Deposit date: | 2024-06-13 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural definition of pseudorabies virus dUTPase reveals a novel folding dimer in the herpesviruses family
To Be Published
|
|
2VHL
 
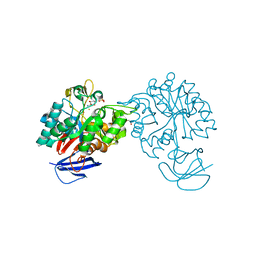 | | The Three-dimensional structure of the N-Acetylglucosamine-6- phosphate deacetylase from Bacillus subtilis | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, FE (III) ION, N-ACETYLGLUCOSAMINE-6-PHOSPHATE DEACETYLASE, ... | | Authors: | Vincent, F, Yates, D, Garman, E, Davies, G.J. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Three-Dimensional Structure of the N-Acetylglucosamine-6-Phosphate Deacetylase from Bacillus Subtilis
J.Biol.Chem., 279, 2004
|
|
8ZYB
 
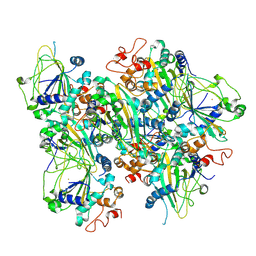 | |
8ZWT
 
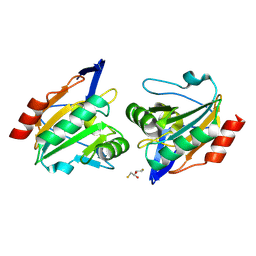 | | Crystal structure of atypical two cysteine thiol peroxidase from Staphylococcus aureus | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Thiol peroxidase | | Authors: | Shukla, M, Maji, S, Yadav, V.K, Bhattacharyya, S. | | Deposit date: | 2024-06-13 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of atypical two cysteine thiol peroxidase from Staphylococcus aureus
To Be Published
|
|
5JQC
 
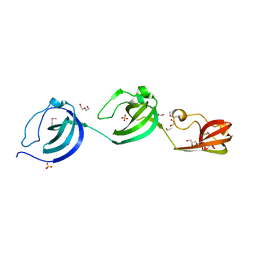 | | Crystal structure putative autolysin from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1076 protein, ... | | Authors: | Chang, C, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal structure putative autolysin from Listeria monocytogenes
To Be Published
|
|
8ZU5
 
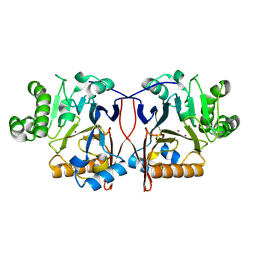 | |
2I2A
 
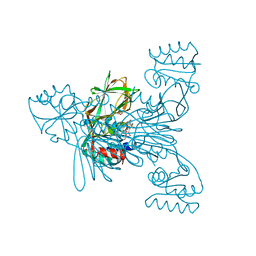 | | Crystal structure of LmNADK1 from Listeria monocytogenes | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Labesse, G. | | Deposit date: | 2006-08-16 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NAD kinases use substrate-assisted catalysis for specific recognition of NAD.
J.Biol.Chem., 282, 2007
|
|
