4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
1RXF
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
1RXT
 
 | | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase. | | Descriptor: | COBALT (II) ION, Glycylpeptide N-tetradecanoyltransferase 1, SULFATE ION | | Authors: | Yang, J, Wang, Y, Frey, G, Abeles, R.H, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase
To be Published
|
|
7Y8D
 
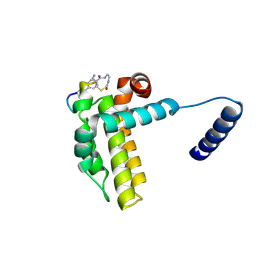 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7BN1
 
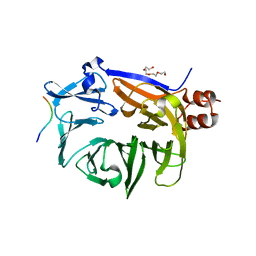 | |
7YA5
 
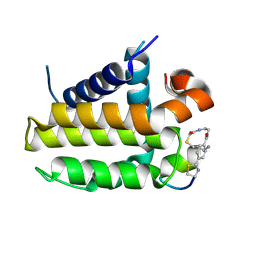 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
8VDS
 
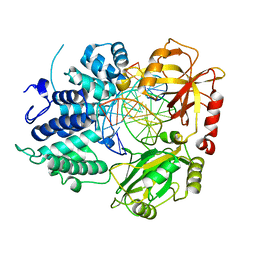 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
8VDT
 
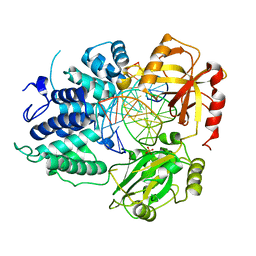 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
4C9K
 
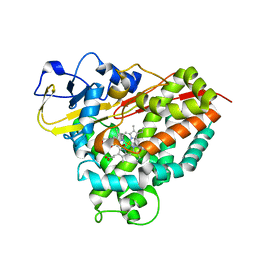 | |
6AMK
 
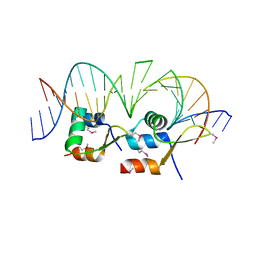 | | Structure of Streptomyces venezuelae BldC-whiI opt complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*TP*TP*AP*CP*CP*CP*GP*AP*AP*TP*TP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*AP*TP*TP*CP*GP*GP*GP*TP*AP*AP*TP*TP*CP*GP*GP*GP*CP*A)-3'), Putative DNA-binding protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-08-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | The MerR-like protein BldC binds DNA direct repeats as cooperative multimers to regulate Streptomyces development.
Nat Commun, 9, 2018
|
|
2G6V
 
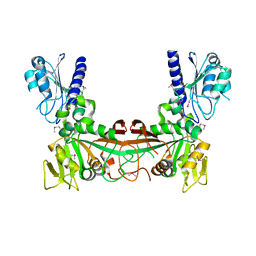 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
2G7K
 
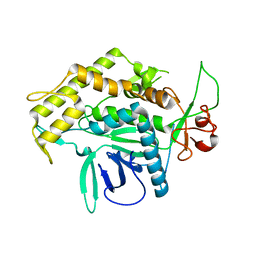 | | Structure of the Light Chain of Botulinum Neurotoxin, Serotype A Bound to small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Crawford, A, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate.
Biochemistry, 45, 2006
|
|
2XXB
 
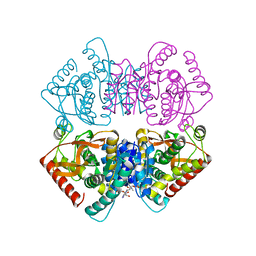 | |
7ONS
 
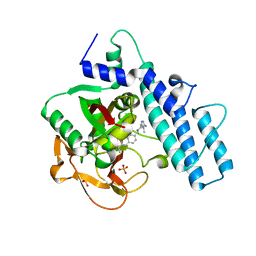 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
5UYZ
 
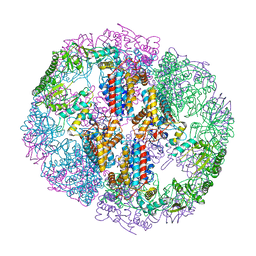 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) mutant His147Arg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
1NVW
 
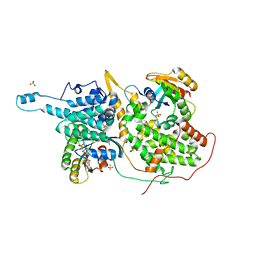 | | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Margarit, S.M, Sondermann, H, Hall, B.E, Nagar, B, Hoelz, A, Pirruccello, M, Bar-Sagi, D, Kuriyan, J. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for feedback activation by RasGTP of the Ras-specific nucleotide exchange factor SOS
Cell(Cambridge,Mass.), 112, 2003
|
|
3DW9
 
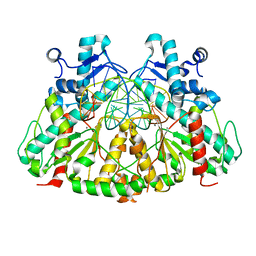 | | SgrAI with cognate DNA and manganese bound | | Descriptor: | DNA (5'-D(*DGP*DAP*DGP*DTP*DCP*DCP*DAP*DCP*DCP*DGP*DGP*DTP*DGP*DGP*DAP*DCP*DTP*DC)-3'), MANGANESE (II) ION, SgraIR restriction enzyme | | Authors: | Dunten, P.W, Horton, N.C, Little, E.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of SgrAI bound to DNA; recognition of an 8 base pair target.
Nucleic Acids Res., 36, 2008
|
|
2Y2X
 
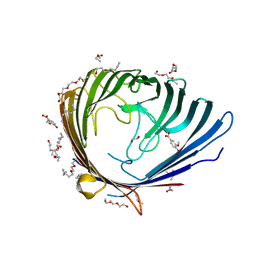 | |
4P15
 
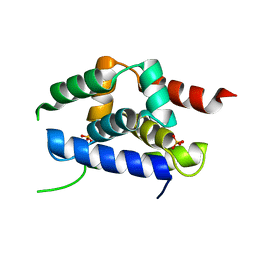 | | Structure of the ClpC N-terminal domain from an alkaliphilic Bacillus lehensis G1 species | | Descriptor: | Bacillus lehensis ClpC N-terminal domain, SULFATE ION | | Authors: | Rashid, S.A, Littler, D.R, Illias, R.M, Murad, A.M.A, Rossjohn, J, Beddoe, T, Mahadi, N.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the ClpC N-terminal domain at 1.85 Angstroms resolution from an alkaliphilic Bacillus lehensis G1 species
To Be Published
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
4EMW
 
 | | Crystal Structure of Staphylococcus aureus bound with the covalent inhibitor EtVC-CoA | | Descriptor: | CHLORIDE ION, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Edwards, J.S, Wallace, B.D, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4P55
 
 | |
