1KGC
 
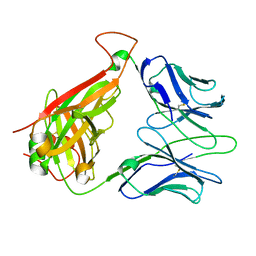 | | Immune Receptor | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Kjer-Nielsen, L, Clements, C.S, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance
STRUCTURE, 10, 2002
|
|
9CYL
 
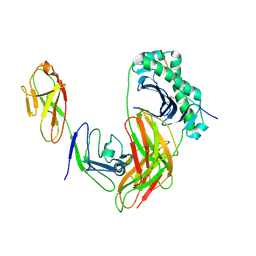 | | Structure of LAG3 loop1 deletion bound to the MHC class II molecule I-A(b) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Class-II-associated invariant chain peptide, H-2 class II histocompatibility antigen, ... | | Authors: | Ming, Q, Tran, T.H, Antfolk, D, Luca, V.C. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.66 Å) | | Cite: | Structural basis for mouse LAG3 interactions with the MHC class II molecule I-A b.
Nat Commun, 15, 2024
|
|
6P95
 
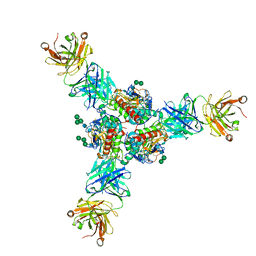 | | Structure of Lassa virus glycoprotein in complex with Fab 25.6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
1KH7
 
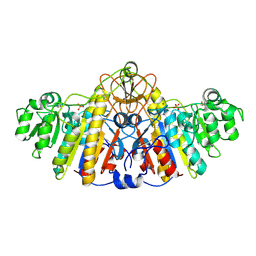 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) | | Descriptor: | MAGNESIUM ION, SULFATE ION, ZINC ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHL
 
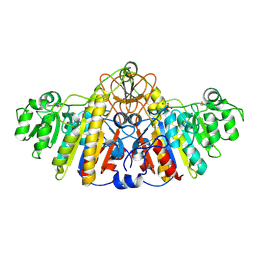 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KXS
 
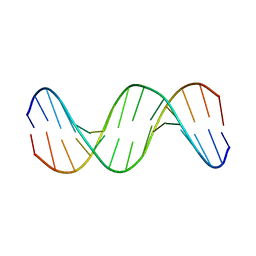 | |
1KFV
 
 | | Crystal Structure of Lactococcus lactis Formamido-pyrimidine DNA Glycosylase (alias Fpg or MutM) Non Covalently Bound to an AP Site Containing DNA. | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*C)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*G)-3', Formamido-pyrimidine DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA.
EMBO J., 21, 2002
|
|
1KI1
 
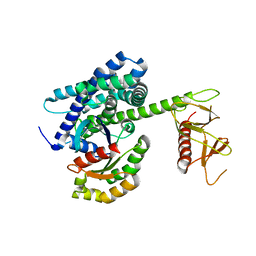 | | Guanine Nucleotide Exchange Region of Intersectin in Complex with Cdc42 | | Descriptor: | G25K GTP-binding protein, placental isoform, SULFATE ION, ... | | Authors: | Snyder, J.T, Pruitt, W.M, Der, C.J, Sondek, J. | | Deposit date: | 2001-12-02 | | Release date: | 2002-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the selective activation of Rho GTPases by Dbl exchange factors.
Nat.Struct.Biol., 9, 2002
|
|
7AB5
 
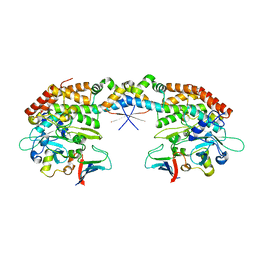 | |
6ZNC
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human wild-type p53DBD bound to DNA and MQ: wt-DNA-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Rozenberg, H, Degtjarik, O, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
1KJV
 
 | | TAP-B-associated rat MHC class I molecule | | Descriptor: | Mature alpha chain of major histocompatibility complex class I antigen (HEAVY CHAIN), SULFATE ION, beta-2-microglobulin, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-05 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
1KKE
 
 | |
7ADI
 
 | | KirBac3.1 W46R: role of a highly conserved tryptophan at the membrane-water interface of Kir channel | | Descriptor: | Inward rectifier potassium channel Kirbac3.1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Venien-Bryan, C, Fagnen, C, De Zorzi, R, Bannwarth, L, Oubella, I, Haouz, A. | | Deposit date: | 2020-09-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Integrative Study of the Structural and Dynamical Properties of a KirBac3.1 Mutant: Functional Implication of a Highly Conserved Tryptophan in the Transmembrane Domain.
Int J Mol Sci, 23, 2021
|
|
1KKP
 
 | | Crystal Structure of Serine Hydroxymethyltransferase complexed with Serine | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, Serine Hydroxymethyltransferase | | Authors: | Trivedi, V, Gupta, A, Jala, V.R, Saravanan, P, Rao, G.S.J, Rao, N.A, Savithri, H.S, Subramanya, H.S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism.
J.Biol.Chem., 277, 2002
|
|
1KG6
 
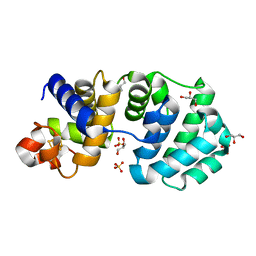 | | Crystal structure of the K142R mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KGN
 
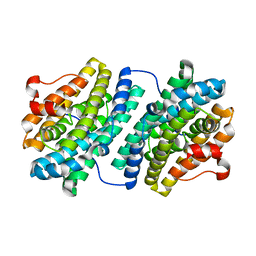 | | R2F from Corynebacterium Ammoniagenes in its oxidised, Fe containing, form | | Descriptor: | FE (III) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
1KHE
 
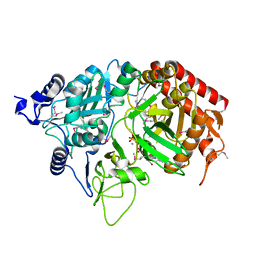 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
7AB4
 
 | |
7A3N
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | CALCIUM ION, Core protein, EDE1 C10 Fab | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
1KHR
 
 | |
1KL3
 
 | |
1KLP
 
 | | The Solution Structure of Acyl Carrier Protein from Mycobacterium tuberculosis | | Descriptor: | MEROMYCOLATE EXTENSION ACYL CARRIER PROTEIN | | Authors: | Wong, H.C, Liu, G, Zhang, Y.-M, Rock, C.O, Zheng, J. | | Deposit date: | 2001-12-12 | | Release date: | 2002-06-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of acyl carrier protein from Mycobacterium tuberculosis.
J.Biol.Chem., 277, 2002
|
|
7AB3
 
 | |
1KMP
 
 | | Crystal structure of the Outer Membrane Transporter FecA Complexed with Ferric Citrate | | Descriptor: | CITRIC ACID, FE (III) ION, IRON(III) DICITRATE TRANSPORT PROTEIN FECA, ... | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KK3
 
 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
