2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
1QXA
 
 | | Crystal structure of Sortase B complexed with Gly3 | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NPQTN specific sortase B, peptide GLY-GLY-GLY | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-05 | | Release date: | 2004-04-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
2F11
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor | | Descriptor: | 2-methylpropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with isobutyl ether mimetic Inhibitor
To be Published
|
|
1MT4
 
 | | Structure of 23S ribosomal RNA hairpin 35 | | Descriptor: | 23S ribosomal Hairpin 35 | | Authors: | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
1MT3
 
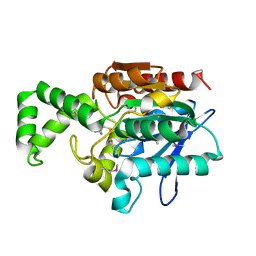 | | Crystal Structure of the Tricorn Interacting Factor Selenomethionine-F1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
1SNT
 
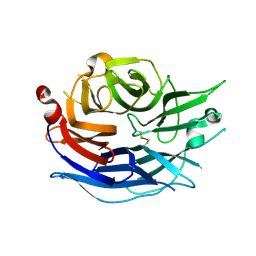 | | Structure of the human cytosolic sialidase Neu2 | | Descriptor: | Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
1IIC
 
 | |
1SP3
 
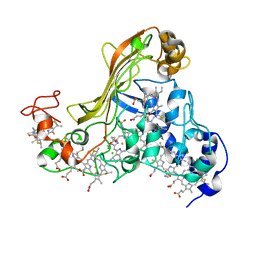 | | Crystal structure of octaheme cytochrome c from Shewanella oneidensis | | Descriptor: | HEME C, THIOCYANATE ION, cytochrome c, ... | | Authors: | Mowat, C.G, Rothery, E, Miles, C.S, McIver, L, Doherty, M.K, Drewette, K, Taylor, P, Walkinshaw, M.D, Chapman, S.K, Reid, G.A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SO7
 
 | | Maltose-induced structure of the human cytolsolic sialidase Neu2 | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
2HZY
 
 | |
2BKD
 
 | | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | | Descriptor: | Fragile X messenger ribonucleoprotein 1 | | Authors: | Ramos, A, Hollingworth, D, Adinolfi, S, Castets, M, Kelly, G, Frenkiel, T.A, Bardoni, B, Pastore, A. | | Deposit date: | 2005-02-15 | | Release date: | 2006-01-18 | | Last modified: | 2023-02-01 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction.
Structure, 14, 2006
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
1T8R
 
 | |
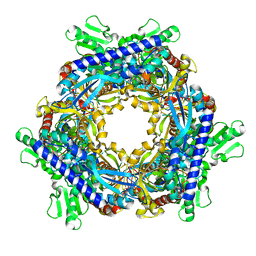
1T8Y
 
 | |
2TSS
 
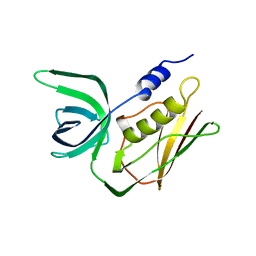 | | TOXIC SHOCK SYNDROME TOXIN-1 FROM STAPHYLOCOCCUS AUREUS: ORTHORHOMBICC222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-04 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
2F12
 
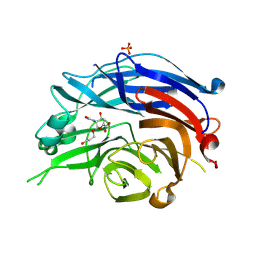 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor | | Descriptor: | 3-hydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 3- hydroxypropyl ether mimetic Inhibitor
To be Published
|
|
1QWZ
 
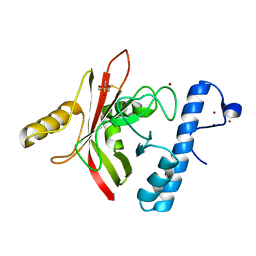 | | Crystal structure of Sortase B from S. aureus complexed with MTSET | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, NICKEL (II) ION, NPQTN specific sortase B, ... | | Authors: | Zong, Y, Mazmanian, S.K, Schneewind, O, Narayana, S.V. | | Deposit date: | 2003-09-03 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall
Structure, 12, 2004
|
|
2F27
 
 | | Crystal Structure of the Human Sialidase Neu2 E111Q-Q112E Double Mutant in Complex with DANA Inhibitor | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 E111Q-Q112E Double Mutant in Complex with DANA Inhibitor
To be Published
|
|
1JAM
 
 | | Crystal structure of apo-form of Z. Mays CK2 protein kinase alpha subunit | | Descriptor: | CASEIN KINASE II, ALPHA CHAIN | | Authors: | Battistutta, R, De Moliner, E, Sarno, S, Zanotti, G, Pinna, L.A. | | Deposit date: | 2001-05-31 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole.
Protein Sci., 10, 2001
|
|
2F28
 
 | | Crystal Structure of the Human Sialidase Neu2 Q116E Mutant | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 Q116E Mutant
To be Published
|
|
1R3Y
 
 | | Uroporphyrinogen Decarboxylase in complex with coproporphyrinogen-III | | Descriptor: | COPROPORPHYRINOGEN III, Uroporphyrinogen Decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Kushner, J.P, Hill, C.P. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase
Embo J., 22, 2003
|
|
1XQY
 
 | | Crystal structure of F1-mutant S105A complex with PRO-LEU-GLY-GLY | | Descriptor: | PLGG, PROLINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XQX
 
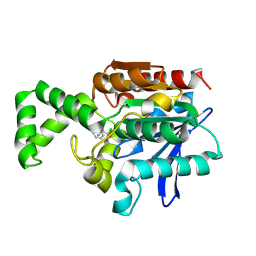 | | Crystal structure of F1-mutant S105A complex with PCK | | Descriptor: | PHENYLALANYLMETHYLCHLORIDE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-13 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1XRM
 
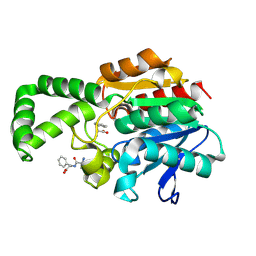 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Ala-Phe | | Descriptor: | ALANINE, PHENYLALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1NM5
 
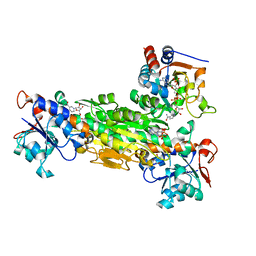 | | R. rubrum transhydrogenase (dI.Q132N)2(dIII)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Van Boxel, G.I, Quirk, P.G, Cotton, N.P, White, S.A, Jackson, J.B. | | Deposit date: | 2003-01-09 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutamine 132 in the NAD(H)-binding component of proton-translocating transhydrogenase tethers the nucleotides before hydride transfer.
Biochemistry, 42, 2003
|
|
