2XCQ
 
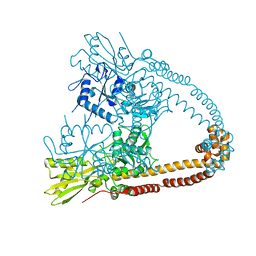 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
9IL7
 
 | | Crystal Structure of SME E166A in Complex with Ceftaroline Fosamil | | Descriptor: | (2~{R})-2-[(1~{R})-1-[[(2~{Z})-2-ethoxyimino-2-[5-(phosphonoamino)-1,2,4-thiadiazol-3-yl]ethanoyl]amino]-2-oxidanylidene-ethyl]-5-[[4-(1-methylpyridin-4-yl)-1,3-thiazol-2-yl]sulfanyl]-3,6-dihydro-2~{H}-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dhankhar, K, Hazra, S. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SME E166A in Complex with Ceftaroline Fosamil
To Be Published
|
|
6R9R
 
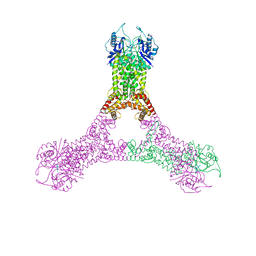 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | Descriptor: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Molina, R, Montoya, G, Sofos, N, Stella, S. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
2IID
 
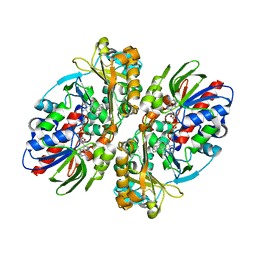 | | Structure of L-amino acid oxidase from Calloselasma rhodostoma in complex with L-phenylalanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase, ... | | Authors: | Moustafa, I.M, Foster, S, Lyubimov, A.Y, Vrielink, A. | | Deposit date: | 2006-09-27 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of LAAO from Calloselasma rhodostoma with an L-Phenylalanine Substrate: Insights into Structure and Mechanism
J.Mol.Biol., 364, 2006
|
|
8EV1
 
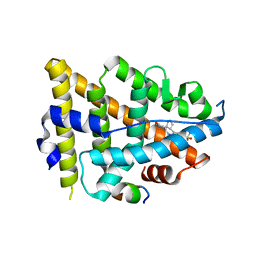 | | Dual Modulators | | Descriptor: | (3aR,4S,9bS)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3aS,4R,9bR)-4-(4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, Estrogen Receptor, ... | | Authors: | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | Deposit date: | 2022-10-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
8EV2
 
 | | Dual Modulators | | Descriptor: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | Authors: | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | Deposit date: | 2022-10-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
2XCS
 
 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCR
 
 | | The 3.5A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase complexed with GSK299423 and DNA | | Descriptor: | 5'-D(*5UA*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
6QOY
 
 | | Crystal structure of L1 protease Lysobacter sp. XL1 in complex with AEBSF | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, 4-(2-azanylethyl)benzenesulfonic acid, CHLORIDE ION, ... | | Authors: | Gabdulkhakov, A, Tishchenko, S, Kudryakova, I, Afoshin, A, Vasilyeva, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serine bacteriolytic protease L1 of Lysobacter sp. XL1 complexed with protease inhibitor AEBSF: features of interaction
Process Biochem, 2019
|
|
2XCO
 
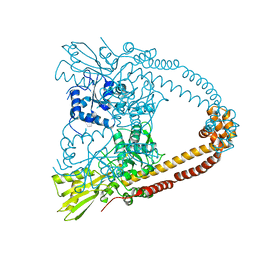 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2JYH
 
 | |
9JOA
 
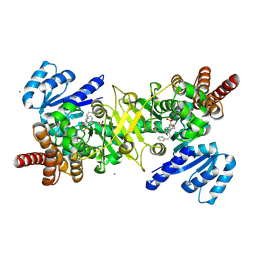 | | PfDXR - Mn2+ - MAMK89 ternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-09-24 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reverse N-Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR)
ACS Infect Dis, 10, 2024
|
|
9JOB
 
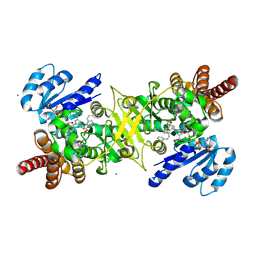 | | PfDXR - Mn2+ - NADPH - MAMK89 quaternary complex | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, CALCIUM ION, ... | | Authors: | Takada, S, Sakamoto, Y, Tanaka, N. | | Deposit date: | 2024-09-24 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Reverse N-Substituted Hydroxamic Acid Derivatives of Fosmidomycin Target a Previously Unknown Subpocket of 1-Deoxy-d-xylulose 5-Phosphate Reductoisomerase (DXR)
ACS Infect Dis, 10, 2024
|
|
2JYF
 
 | |
2Y7Z
 
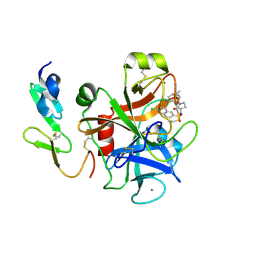 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-[(1S)-1-DIMETHYLAMINO-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y81
 
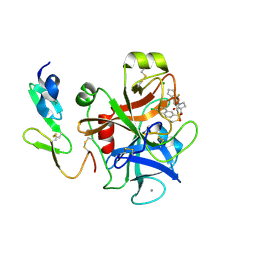 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2R)-2--PYRROLIDINYL] PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2M16
 
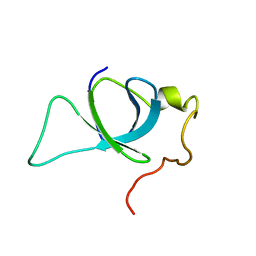 | | P75/LEDGF PWWP Domain | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2012-11-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes.
Nucleic Acids Res., 41, 2013
|
|
6QZQ
 
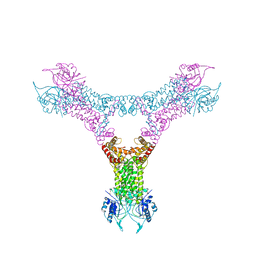 | |
2Y82
 
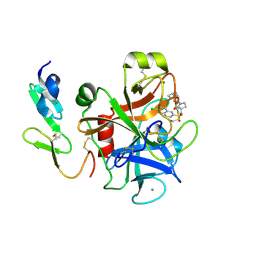 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2S)-2-PYRROLIDINYL]PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
9GZT
 
 | | WT-IAPP cryo-EM structure Type LLUU - control reaction | | Descriptor: | Islet amyloid polypeptide | | Authors: | Wilkinson, M, Taylor, A.I.P, Xu, Y, Chakraborty, P, Brinkworth, A, Willis, L.F, Zhuravleva, A, Ranson, N.A, Foster, R, Radford, S.E. | | Deposit date: | 2024-10-04 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Kinetic Steering of Amyloid Formation and Polymorphism by Canagliflozin, a Type-2 Diabetes Drug.
J.Am.Chem.Soc., 147, 2025
|
|
2Y80
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-(DIMETHYLAMINO)-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2WC9
 
 | | Crystal structure of the g2p (large terminase) nuclease domain from the bacteriophage SPP1 with bound Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TERMINASE LARGE SUBUNIT | | Authors: | Smits, C, Chechik, M, Kovalevskiy, O.V, Shevtsov, M.B, Foster, A.W, Alonso, J.C, Antson, A.A. | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Nuclease Activity of a Bacteriophage Large Terminase.
Embo Rep., 10, 2009
|
|
4JYG
 
 | | Crystal structure of RARbeta LBD in complex with agonist BMS411 [4-{[(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)carbonyl]amino}benzoic acid] | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-{[(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)carbonyl]amino}benzoic acid, CITRATE ANION, ... | | Authors: | Nadendla, E.K, Teyssier, C, Germain, P, Delfosse, V, Bourguet, W. | | Deposit date: | 2013-03-29 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An Unexpected Mode Of Binding Defines BMS948 as A Full Retinoic Acid Receptor beta (RAR beta , NR1B2) Selective Agonist.
Plos One, 10, 2015
|
|
2N3K
 
 | | Human Brd4 ET domain in complex with MLV Integrase C-term | | Descriptor: | Bromodomain-containing protein 4, MLV integrase | | Authors: | Crowe, B.L, Foster, M.P. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3OO0
 
 | | Structure of apo CheY A113P | | Descriptor: | AMMONIUM ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Guanga, G.P, Miller, P.J, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-31 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Role of Position K+4 in the Phosphorylation and Dephosphorylation Reaction Kinetics of the CheY Response Regulator.
Biochemistry, 60, 2021
|
|
