5NP0
 
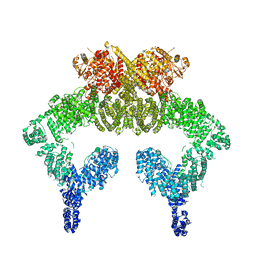 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NJI
 
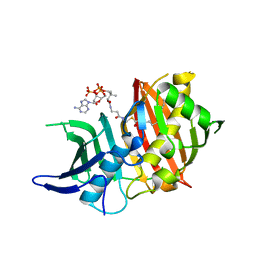 | | Structure of the dehydratase domain of PpsC from Mycobacterium tuberculosis in complex with C12:1-CoA | | Descriptor: | Phthiocerol/phenolphthiocerol synthesis polyketide synthase type I PpsC, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (~{E})-dodec-2-enethioate | | Authors: | Gavalda, S, Faille, A, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into Substrate Modification by Dehydratases from Type I Polyketide Synthases.
J. Mol. Biol., 429, 2017
|
|
5NJM
 
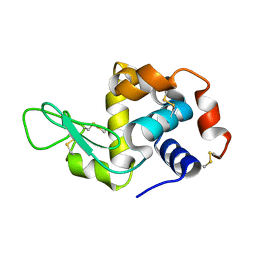 | | Lysozyme room-temperature structure determined by serial millisecond crystallography | | Descriptor: | Lysozyme C | | Authors: | Weinert, T, Vera, L, Marsh, M, James, D, Gashi, D, Nogly, P, Jaeger, K, Standfuss, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NQT
 
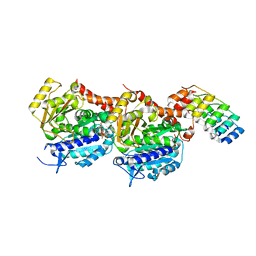 | | Tubulin Darpin room-temperature structure determined by serial millisecond crystallography | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
7KVV
 
 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, Squash RNA aptamer bound to DFHO | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVT
 
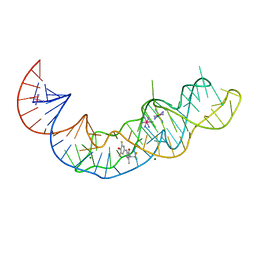 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T with iridium (III) ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, IRIDIUM HEXAMMINE ION, MAGNESIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
7KVU
 
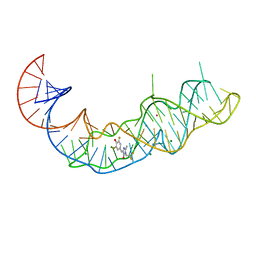 | | Crystal structure of Squash RNA aptamer in complex with DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Truong, L, Ferre-D'Amare, A.R. | | Deposit date: | 2020-11-28 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold.
Nat.Chem.Biol., 18, 2022
|
|
5O4U
 
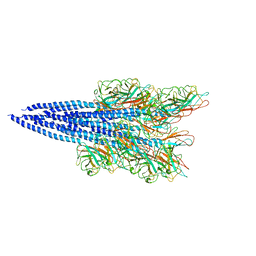 | | The flagellin of Pyrococcus furiosus | | Descriptor: | Flagellin | | Authors: | Daum, B, Vonck, J. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure andin situorganisation of thePyrococcus furiosusarchaellum machinery.
Elife, 6, 2017
|
|
2GVU
 
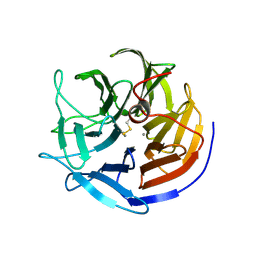 | |
2GVV
 
 | |
5OVB
 
 | | Crystal structure of human BRD4(1) bromodomain in complex with DR46 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-08-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
2GVW
 
 | |
7L6C
 
 | |
2KIR
 
 | | Solution structure of a designer toxin, mokatoxin-1 | | Descriptor: | Designer toxin | | Authors: | Biancalana, M, Koide, A, Takacs, Z, Goldstein, S, Koide, S. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A designer ligand specific for Kv1.3 channels from a scorpion neurotoxin-based library.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7KSL
 
 | | Substrate-free human mitochondrial LONP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
7MKE
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKI
 
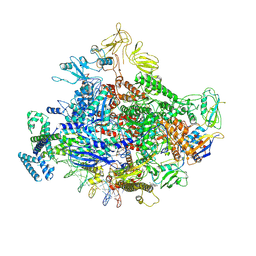 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
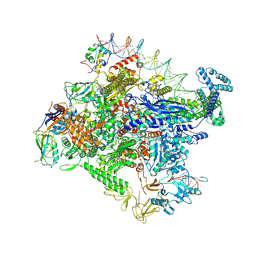 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7M2K
 
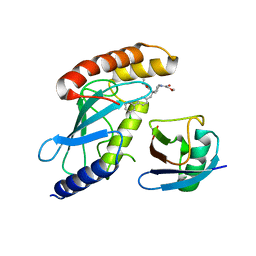 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
5OL0
 
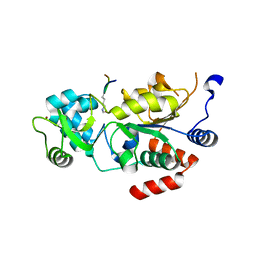 | | Structure of Leishmania infantum Silent Information Regulator 2 related protein 1 (LiSIR2rp1) in complex with acetylated p53 peptide | | Descriptor: | Cellular tumor antigen p53, Putative silent information regulator 2,Putative silent information regulator 2, ZINC ION | | Authors: | Ronin, C, Ciesielski, F, Ciapetti, P. | | Deposit date: | 2017-07-26 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the Leishmania infantum Silent Information Regulator 2 related protein 1: Implications to protein function and drug design.
PLoS ONE, 13, 2018
|
|
2GVX
 
 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
2HS4
 
 | | T. maritima PurL complexed with FGAR and AMPPCP | | Descriptor: | MAGNESIUM ION, N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
