7FBV
 
 | |
5IOH
 
 | | RepoMan-PP1a (protein phosphatase 1, alpha isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
2K8T
 
 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6R,8S,11R)-configuration opposite dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
5Z1C
 
 | | The crystal structure of uPA in complex with 4-Iodobenzylamine at pH7.4 | | Descriptor: | 1-(4-iodophenyl)methanamine, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Zhang, X, Luo, Z.P, Huang, M.D. | | Deposit date: | 2017-12-25 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 49, 2018
|
|
3GBD
 
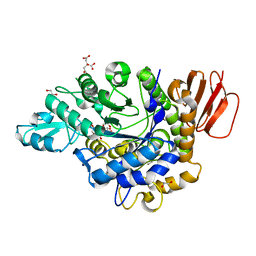 | | Crystal structure of the isomaltulose synthase SmuA from Protaminobacter rubrum | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Sucrose isomerase SmuA from Protaminobacter rubrum | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural determinants of product specificity of sucrose isomerases
Febs Lett., 583, 2009
|
|
1EVB
 
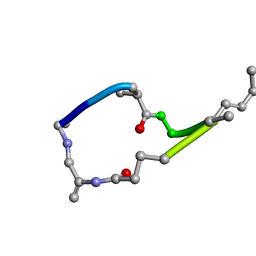 | |
1EVA
 
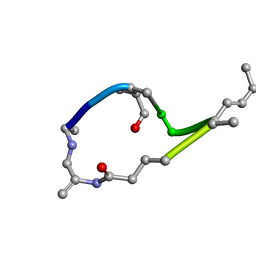 | |
1EVD
 
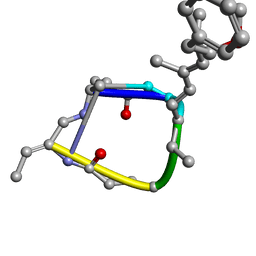 | |
4QOA
 
 | |
9N77
 
 | | SSU processome maturation and disassembly, State K | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
9QFQ
 
 | | Cryo-EM structure of the cofilactin barbed end bound by AIP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-12 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
4QND
 
 | | Crystal structure of a SemiSWEET | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Chemical transport protein, ... | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
5M33
 
 | | Structural tuning of CD81LEL (space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
9NZM
 
 | | Crystal Structure of Kirsten Rat Sarcoma G12C Complexed with GMPPNP and Covalently Bound to an Adduct of {(2S)-4-[7-(8-chloronaphthalen-1-yl)-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]-1-[(2Z)-2-fluoro-3-(pyridin-2-yl)prop-2-enoyl]piperazin-2-yl}acetonitrile | | Descriptor: | Isoform 2B of GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Sheriff, S, Pokross, M, Witmer, M. | | Deposit date: | 2025-04-01 | | Release date: | 2025-11-05 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Novel covalent warhead imparts potent activity against both GDP- and GTP-bound forms of KRAS G12C
To be published
|
|
5ZNJ
 
 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
8RG1
 
 | | Soluble glucose dehydrogenase from acinetobacter calcoaceticus - wild type pH8 | | Descriptor: | 3-(3,5-dicarboxy-1~{H}-pyrrol-2-yl)pyridine-2,4,6-tricarboxylic acid, CALCIUM ION, LITHIUM ION, ... | | Authors: | Lublin, V, Chavas, L, Stines-Chaumeil, C, Kauffmann, B, Giraud, M.F, Thompson, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Does Acinetobacter calcoaceticus glucose dehydrogenase produce self-damaging H2O2?
Biosci.Rep., 44, 2024
|
|
6W9U
 
 | | Structure of human MAIT A-F7 TCR in complex with patient MR1-R9H-Ac-6-FP | | Descriptor: | 1-[[6-(1-$l^{1}-oxidanylethyl)-4-$l^{3}-oxidanylidene-2,3,6,8~{a}-tetrahydropteridin-2-yl]-$l^{2}-azanyl]ethanone, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1 .
Sci Immunol, 5, 2020
|
|
5M5U
 
 | |
9N75
 
 | | SSU processome maturation and disassembly, State I | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S11-A, ... | | Authors: | Buzovetsky, O, Klinge, S. | | Deposit date: | 2025-02-05 | | Release date: | 2025-11-05 | | Last modified: | 2025-11-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Helicase-mediated mechanism of SSU processome maturation and disassembly.
Nature, 2025
|
|
4QPV
 
 | |
9R0H
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with compound 6 (ORIC-533) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Moore, J.T, Ivic, N, Lammens, A, Krapp, S, Maskos, K. | | Deposit date: | 2025-04-24 | | Release date: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of ORIC-533, an Orally Bioavailable CD73 Inhibitor That Maintains Activity in High AMP Environments to Reverse Tumor Immunosuppression.
J.Med.Chem., 2025
|
|
6SXI
 
 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
4YGT
 
 | |
8VDO
 
 | |
