6TQI
 
 | | I-MOTIF STRUCTURE FORMED FROM THE C STRAND OF A HUMAN TELOMERE FRAGMENT | | Descriptor: | DNA (5'-*TP*AP*AP*CP*CP*CP*TP*AP*A-3') | | Authors: | Parkinson, G.N, Wagner, A, Viladoms-Claverol, J, Duman, R, El-Omari, K. | | Deposit date: | 2019-12-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
4C11
 
 | |
6F57
 
 | |
6JJI
 
 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 (1:1) | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJH
 
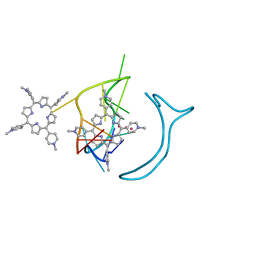 | | Crystal structure of a two-quartet RNA parallel G-quadruplex complexed with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, POTASSIUM ION, RNA (5'-R(*GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3') | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
6JJF
 
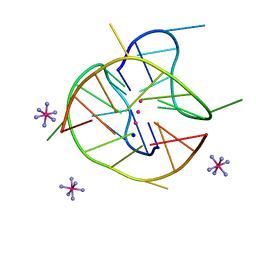 | | Crystal structure of a two-quartet DNA mixed-parallel/antiparallel G-quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*GP*CP*TP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A)-3'), POTASSIUM ION, ... | | Authors: | Zhang, Y.S, EI Omari, K, Duman, R, Wagner, A, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths.
Nucleic Acids Res., 48, 2020
|
|
3DQS
 
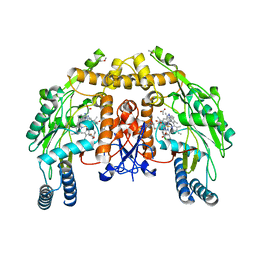 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3B3P
 
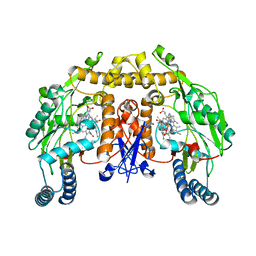 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQR
 
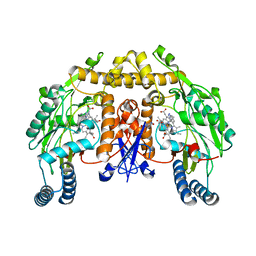 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3DQT
 
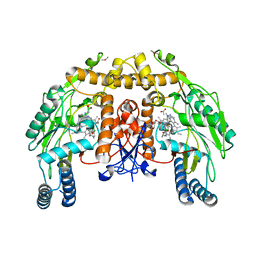 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{trans-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3B3O
 
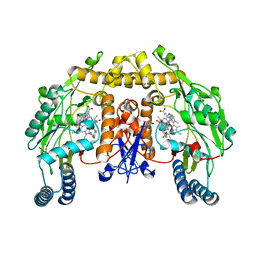 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-AMINO-4-METHYLPYRIDIN-2-YL)METHYL]PYRROLIDIN-3-YL}-N'-(4-CHLOROBENZYL)ETHANE-1,2-DIAMINE, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
5KB2
 
 | | Crystal Structure of a Tris-thiolate Zn(II)S3O Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | ZINC ION, Zn(II)(H2O)(GRAND Coil Ser-L12AL16C)3- | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5KB0
 
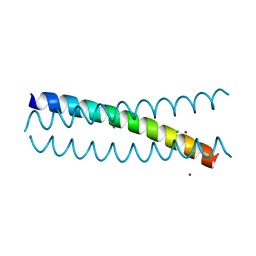 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
8UZL
 
 | | Designed Transmembrane beta-barrel- TMB10_163 | | Descriptor: | Designed Transmembrane beta-barrel TMB10_163, HEXANE-1,6-DIOL | | Authors: | Bera, A.K, Lemma, S.B, Kang, A, Baker, D. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sculpting conducting nanopore size and shape through de novo protein design.
Science, 385, 2024
|
|
5U9T
 
 | | The Tris-thiolate Zn(II)S3Cl Binding Site Engineered by D-Cysteine Ligands in de Novo Three-stranded Coiled Coil Environment | | Descriptor: | ACETATE ION, CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Ruckthong, L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | d-Cysteine Ligands Control Metal Geometries within De Novo Designed Three-Stranded Coiled Coils.
Chemistry, 23, 2017
|
|
5EOJ
 
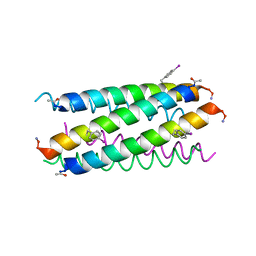 | |
4JXI
 
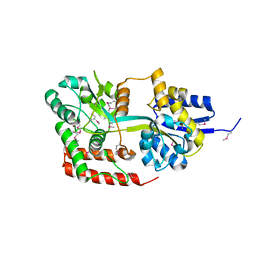 | | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, BROMIDE ION, GLYCEROL, ... | | Authors: | Kuzin, A, Smith, M.D, Richter, F, Lew, S, Seetharaman, R, Bryan, C, Lech, Z, Kiss, G, Moretti, R, Maglaqui, M, Xiao, R, Kohan, E, Smith, M, Everett, J.K, Nguyen, R, Pande, V, Hilvert, D, Kornhaber, G, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Directed evolution and rational design of a de novo designed esterase toward improved catalysis. Northeast Structural Genomics Consortium (NESG) Target OR184
To be Published
|
|
5KB1
 
 | | Crystal Structure of a Tris-thiolate Hg(II) Complex in a de Novo Three Stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, Hg(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, MERCURY (II) ION, ... | | Authors: | Ruckcthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5TPH
 
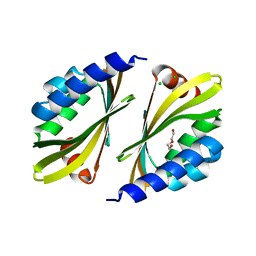 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5K92
 
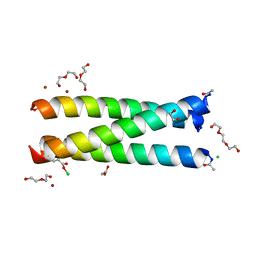 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
8E55
 
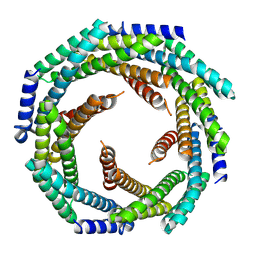 | |
5U35
 
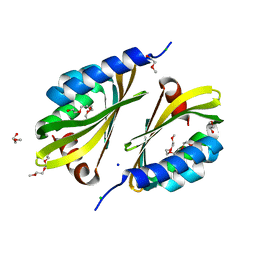 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
7BPP
 
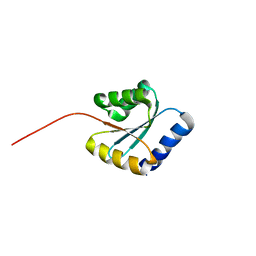 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | Descriptor: | NF5 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPN
 
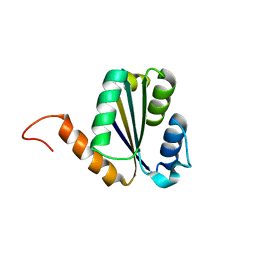 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
