7UJX
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-03-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1A7M
 
 | | LEUKAEMIA INHIBITORY FACTOR CHIMERA (MH35-LIF), NMR, 20 STRUCTURES | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Hinds, M.G, Maurer, T, Zhang, J.-G, Nicola, N.A, Norton, R.S. | | Deposit date: | 1998-03-16 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leukemia inhibitory factor.
J.Biol.Chem., 273, 1998
|
|
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1ZQ3
 
 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
7L1G
 
 | | PRMT5-MEP50 Complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Development of a Flexible and Robust Synthesis of Tetrahydrofuro[3,4- b ]furan Nucleoside Analogues.
J.Org.Chem., 86, 2021
|
|
7PSU
 
 | | Structure of protein kinase CK2alpha mutant K198R associated with the Okur-Chung Neurodevelopmental Syndrome | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Werner, C, Gast, A, Lindenblatt, D, Nickelsen, K, Niefind, K, Jose, J, Hochscherf, J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Enzymological Evidence for an Altered Substrate Specificity in Okur-Chung Neurodevelopmental Syndrome Mutant CK2 alpha Lys198Arg.
Front Mol Biosci, 9, 2022
|
|
4NFT
 
 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
3QWW
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the methyltransferase inhibitor sinefungin | | Descriptor: | SET and MYND domain-containing protein 2, SINEFUNGIN, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
3QWV
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the cofactor product AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 2, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
9ATK
 
 | | BIFUNCTIONAL INHIBITION OF NEUTROPHIL ELASTASE AND CATHEPSIN G by Eap4 of S. aureus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin-G, ... | | Authors: | Mishra, N.B, Herdendorf, T.J, Geisbrecht, B.V. | | Deposit date: | 2024-02-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J.Biol.Chem., 300, 2024
|
|
9ASX
 
 | | BIFUNCTIONAL INHIBITION OF NEUTROPHIL ELASTASE AND CATHEPSIN G by Eap3 of S. aureus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin-G, ... | | Authors: | Mishra, N.B, Herdendorf, T.J, Geisbrecht, B.V. | | Deposit date: | 2024-02-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | S. aureus Eap is a polyvalent inhibitor of neutrophil serine proteases.
J.Biol.Chem., 300, 2024
|
|
5MND
 
 | |
5MXO
 
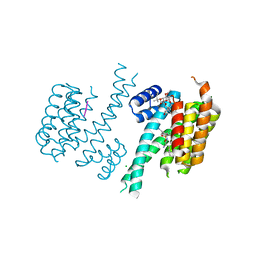 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide stabilized by Fusicoccin-A | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, FUSICOCCIN, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
4L0Y
 
 | |
4L18
 
 | |
2MEV
 
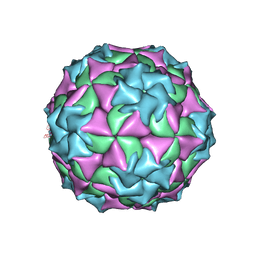 | | STRUCTURAL REFINEMENT AND ANALYSIS OF MENGO VIRUS | | Descriptor: | MENGO VIRUS COAT PROTEIN (SUBUNIT VP1), MENGO VIRUS COAT PROTEIN (SUBUNIT VP2), MENGO VIRUS COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Rossmann, M.G. | | Deposit date: | 1989-04-21 | | Release date: | 1990-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural refinement and analysis of Mengo virus.
J.Mol.Biol., 211, 1990
|
|
4I5H
 
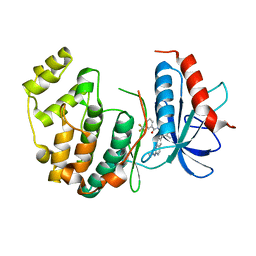 | |
4KXP
 
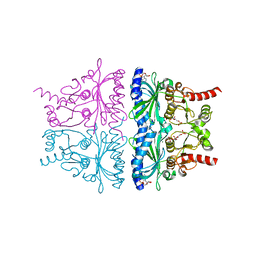 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant I10D in T-state | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2013-05-27 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Displacement of a Catalytically Essential Loop from the Active Site of Mammalian Fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
4L0Z
 
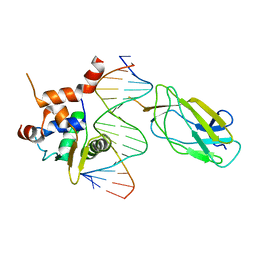 | |
2MPL
 
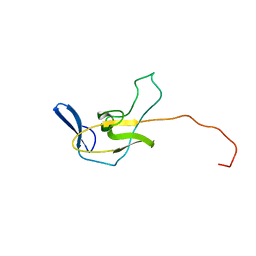 | |
3WU1
 
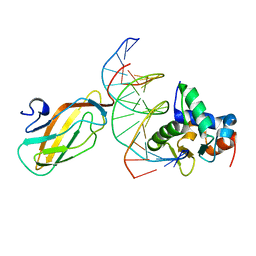 | | Crystal structure of the ETS1-RUNX1-DNA ternary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*T)-3'), Protein C-ets-1, ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel allosteric mechanism on protein-DNA interactions underlying the phosphorylation-dependent regulation of Ets1 target gene expressions.
J.Mol.Biol., 427, 2015
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
8UFZ
 
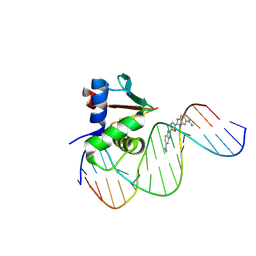 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGCGGAAGTG) in Ternary Complex with DB1976 | | Descriptor: | (2M,2'M)-2,2'-(selenophene-2,5-diyl)di(1H-benzimidazole-6-carboximidamide), DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*TP*TP*AP*T)-3'), ... | | Authors: | Terrell, J.R, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-10-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural studies on the PU.1 inhibitory mechanism by diamidine minor groove binders
To Be Published
|
|
2Z9J
 
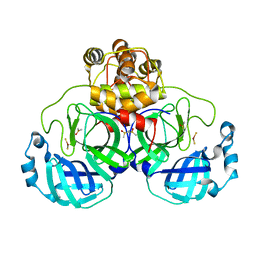 | | Complex structure of SARS-CoV 3C-like protease with EPDTC | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
7A1O
 
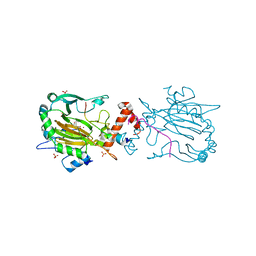 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 4-ethyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 4-ethyl-2-oxoglutarate, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
