2M60
 
 | | Enterocin 7B | | Descriptor: | Enterocin JSB | | Authors: | Lohans, C.T, Towle, K.M, Miskolzie, M, McKay, R.T, van Belkum, M.J, McMullen, L.M, Vederas, J.C. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Linear Leaderless Bacteriocins Enterocin 7A and 7B Resemble Carnocyclin A, a Circular Antimicrobial Peptide
Biochemistry, 52, 2013
|
|
2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
2M62
 
 | |
2M63
 
 | | The protease-resistant N-terminal domain of TIR-domain containing adaptor molecule-1, TICAM-1 | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Kumeta, H, Enokizono, Y, Sakakibara, H, Ogura, K, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of TIR domain-containing adaptor molecule-1, TICAM-1.
J.Biomol.Nmr, 58, 2014
|
|
2M64
 
 | | 1H, 13C and 15N Chemical Shift Assignments for Phl p 5a | | Descriptor: | Phlp5 | | Authors: | Goebl, C, Focke, M, Schrank, E, Madl, T, Kosol, S, Madritsch, C, Flicker, S, Valenta, R, Zangger, K, Tjandra, N. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexible IgE epitope-containing domains of Phl p 5 cause high allergenic activity.
J. Allergy Clin. Immunol., 140, 2017
|
|
2M65
 
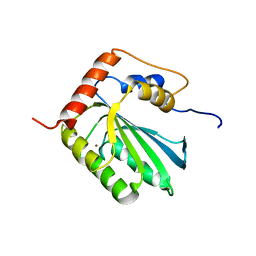 | | NMR structure of human restriction factor APOBEC3A | | Descriptor: | Probable DNA dC->dU-editing enzyme APOBEC-3A, ZINC ION | | Authors: | Byeon, I.L, Byeon, C, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity.
Nat Commun, 4, 2013
|
|
2M66
 
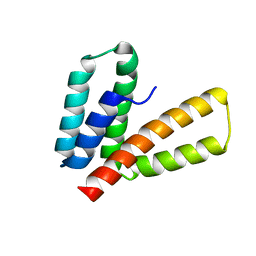 | | Endoplasmic reticulum protein 29 (ERp29) C-terminal domain: 3D Protein Fold Determination from Backbone Amide Pseudocontact Shifts Generated by Lanthanide Tags at Multiple Sites | | Descriptor: | Endoplasmic reticulum resident protein 29 | | Authors: | Yagi, H, Pilla, K, Maleckis, A, Graham, B, Huber, T, Otting, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites
Structure, 21, 2013
|
|
2M67
 
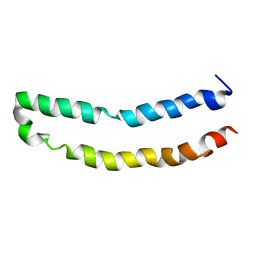 | | Full-length mercury transporter protein MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Lu, G.J, Tian, Y, Vora, N, Marassi, F.M, Opella, S.J. | | Deposit date: | 2013-03-27 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The Structure of the Mercury Transporter MerF in Phospholipid Bilayers: A Large Conformational Rearrangement Results from N-Terminal Truncation.
J.Am.Chem.Soc., 135, 2013
|
|
2M68
 
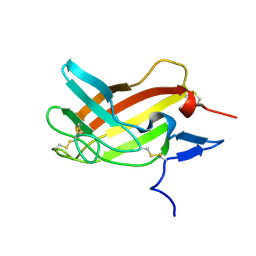 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R in complex with IGF2 (domain 11 structure only) | | Descriptor: | Insulin-like growth factor 2 receptor variant | | Authors: | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | Deposit date: | 2013-03-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2M6A
 
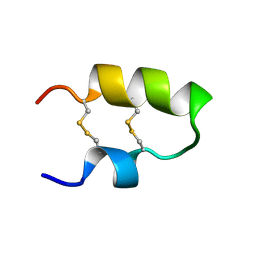 | | NMR spatial structure of the antimicrobial peptide Tk-Amp-X2 | | Descriptor: | Predicted protein | | Authors: | Usmanova, D.R, Mineev, K.S, Arseniev, A.S, Berkut, A.A, Oparin, P.B, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design
J.Biol.Chem., 289, 2014
|
|
2M6B
 
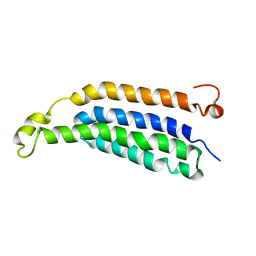 | | Structure of full-length transmembrane domains of human glycine receptor alpha1 monomer subunit | | Descriptor: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | Authors: | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | Deposit date: | 2013-03-28 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
2M6C
 
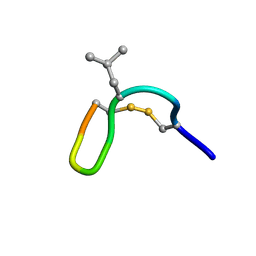 | | NMR solution structure of cis (minor) form of In936 in water | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-28 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6D
 
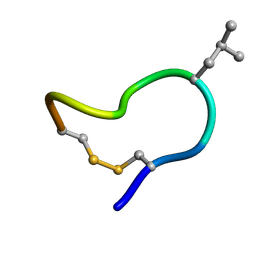 | | NMR solution structure of trans (major) form of In936 in water | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-28 | | Release date: | 2013-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6E
 
 | | NMR solution structure of cis (minor) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6F
 
 | | NMR solution structure of trans (major) form of In936 in Methanol | | Descriptor: | Contryphan-In | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2022-08-24 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6G
 
 | | Solution structure of cis(C2-P3) trans (D5-P6) form of lO959 in water | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6H
 
 | | Solution structure of trans(C2-P3) trans (D5-P6) of LO959 in methanol | | Descriptor: | Contryphan-Lo | | Authors: | Sonti, R. | | Deposit date: | 2013-03-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Conformational diversity in contryphans from Conus venom: cis-trans isomerisation and aromatic/proline interactions in the 23-membered ring of a 7-residue peptide disulfide loop.
Chemistry, 19, 2013
|
|
2M6I
 
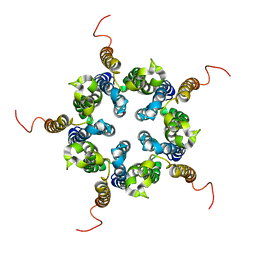 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | Descriptor: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | Authors: | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | Deposit date: | 2013-03-29 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
2M6J
 
 | |
2M6K
 
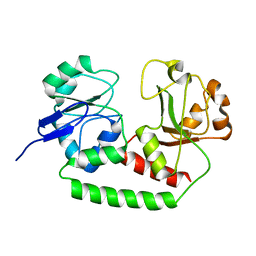 | | Solution structure of the Escherichia coli apo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
2M6L
 
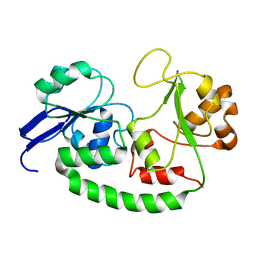 | | Solution structure of the Escherichia coli holo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
2M6M
 
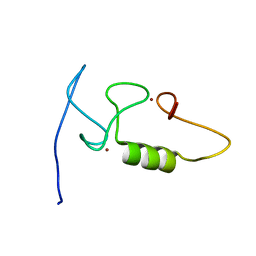 | |
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2M6O
 
 | | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase | | Descriptor: | Uncharacterized protein | | Authors: | Liu, B, Tabib-Salazar, A, Doughty, P, Lewis, R, Ghosh, S, Parsy, M, Simpson, P, Matthews, S, Paget, M. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase.
Nucleic Acids Res., 41, 2013
|
|
2M6P
 
 | |
