8CIE
 
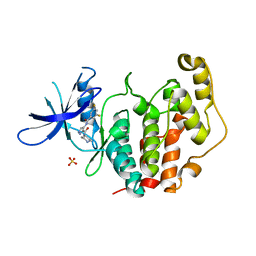 | | Crystal structure of the human CDKL5 kinase domain with compound YL-354 | | Descriptor: | 4-[[3,5-bis(fluoranyl)phenyl]carbonylamino]-~{N}-piperidin-4-yl-1~{H}-pyrazole-3-carboxamide, Cyclin-dependent kinase-like 5, SULFATE ION | | Authors: | Richardson, W, Chen, X, Newman, J.A, Bakshi, S, Lakshminarayana, B, Brooke, L, Bullock, A.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective CDKL5/GSK3 Chemical Probe That Is Neuroprotective.
Acs Chem Neurosci, 14, 2023
|
|
6MPV
 
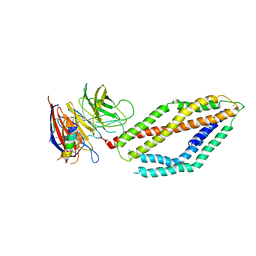 | |
4Z0C
 
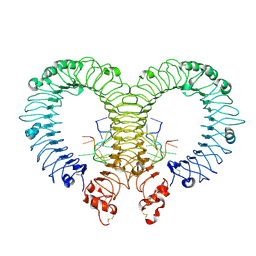 | | Crystal structure of TLR13-ssRNA13 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-R(P*AP*CP*GP*GP*AP*AP*AP*GP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2015-03-26 | | Release date: | 2015-10-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13
Nat.Struct.Mol.Biol., 22, 2015
|
|
5HYX
 
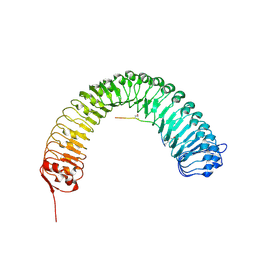 | | Plant peptide hormone receptor RGFR1 in complex with RGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-SER-ASN-PRO-GLY-HIS-HIS-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth
Cell Res., 26, 2016
|
|
8A8E
 
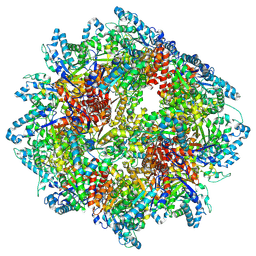 | | PPSA C terminal octahedral structure | | Descriptor: | Phosphoenolpyruvate synthase | | Authors: | Song, W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Survival strategies in the heat Lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermophilic PEP-synthase
To Be Published
|
|
7XOZ
 
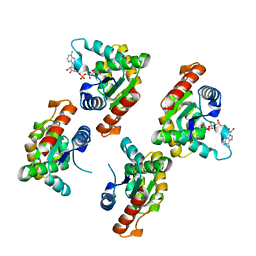 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
5HZ0
 
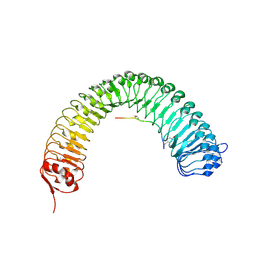 | | Plant peptide hormone receptor RGFR1 in complex with RGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-LYS-PRO-ARG-HIS-HIS-PRO-HYP-ARG-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HZ1
 
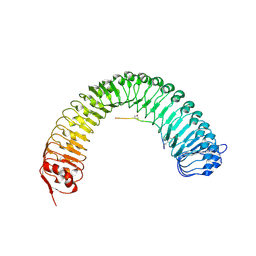 | | Plant peptide hormone receptor RGFR1 in complex with RGF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-ARG-ALA-LYS-HIS-HIS-PRO-HYP-LYS-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HZ3
 
 | | Plant peptide hormone receptor RGFR1 in complex with RGFR5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-PRO-LYS-PRO-SER-THR-ARG-PRO-HYP-ARG-HIS-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Plant Receptor
To Be Published
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
3KD3
 
 | | Crystal structure of a phosphoserine phosphohydrolase-like protein from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphoserine phosphohydrolase-like protein | | Authors: | Nocek, B, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a phosphoserine phosphohydrolase-like protein from Francisella tularensis subsp. tularensis SCHUS4
TO BE PUBLISHED
|
|
3RRO
 
 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, 3-ketoacyl-(acyl-carrier-protein) reductase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
2X1N
 
 | | Truncation and Optimisation of Peptide Inhibitors of CDK2, Cyclin A Through Structure Guided Design | | Descriptor: | 2-METHYL-N-[(1Z)-3-NITROCYCLOHEXA-2,4-DIEN-1-YLIDENE]-4,5-DIHYDRO[1,3]THIAZOLO[4,5-H]QUINAZOLIN-8-AMINE, ACE-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, ... | | Authors: | Kontopidis, G, Andrews, M.J, McInnes, C, Plater, A, Innes, L, Renachowski, S, Cowan, A, Fischer, P.M, McIntyre, N.A, Griffiths, G, Barnett, A.L, Slawin, A.M.Z, Jackson, W, Thomas, M, Zheleva, D.I, Wang, S, Blake, D.G, Westwood, N.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Synthesis, and Evaluation of 2-Methyl- and 2-Amino-N-Aryl-4,5-Dihydrothiazolo[4,5-H]Quinazolin-8-Amines as Ring-Constrained 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Cyclin-Dependent Kinase Inhibitors.
J.Med.Chem., 53, 2010
|
|
9FGO
 
 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
1GYX
 
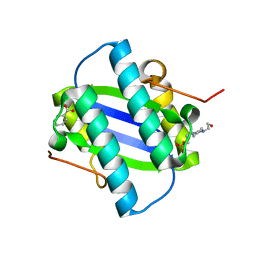 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BENZOIC ACID, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYJ
 
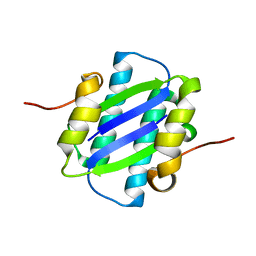 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-23 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
1GYY
 
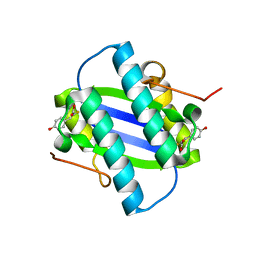 | | The Crystal Structure of YdcE, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia coli, Confirms the Structural Basis for Oligomer Diversity | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, HYPOTHETICAL PROTEIN YDCE | | Authors: | Almrud, J, Kern, A, Wang, S, Czerwinski, R, Johnson, W, Murzin, A, Hackert, M, Whitman, C. | | Deposit date: | 2002-04-30 | | Release date: | 2002-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Crystal Structure of Ydce, a 4-Oxalocrotonate Tautomerase Homologue from Escherichia Coli, Confirms the Structural Basis for Oligomer Diversity
Biochemistry, 41, 2002
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | Descriptor: | Copper-ion-binding protein | | Authors: | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-16 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | To be published
To be Published
|
|
5XLR
 
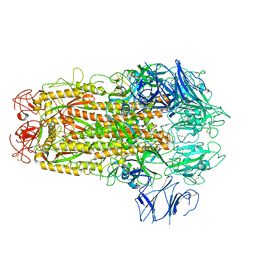 | | Structure of SARS-CoV spike glycoprotein | | Descriptor: | Spike glycoprotein | | Authors: | Gui, M, Song, W, Xiang, Y, Wang, X. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-07 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
2M6M
 
 | |
2LJ2
 
 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
3KWO
 
 | | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni
To be Published
|
|
3LXY
 
 | | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92 | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 4-hydroxythreonine-4-phosphate dehydrogenase from Yersinia pestis CO92
To be Published
|
|
3LUS
 
 | | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | L-CAPTOPRIL, Organic hydroperoxide resistance protein | | Authors: | Nocek, B, Maltseva, N, Makowska-Grzyska, M, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-18 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961
TO BE PUBLISHED
|
|
3M34
 
 | | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-08 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of transketolase in complex with thiamin diphosphate and calcium ion
To be Published
|
|
