8JC8
 
 | | Cryo-EM structure of the LH1 complex from thermochromatium tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, LH1 alpha polypeptide, ... | | Authors: | Wang, G.-L, Yan, Y.-H, Yu, L.-J. | | Deposit date: | 2023-05-10 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Molecular structure and characterization of the Thermochromatium tepidum light-harvesting 1 photocomplex produced in a foreign host.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
6NTC
 
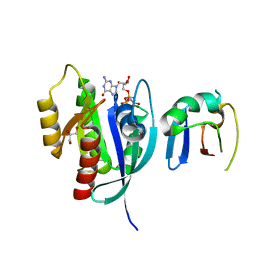 | | Crystal Structure of G12V HRas-GppNHp bound in complex with the engineered RBD variant 1 of CRAF Kinase protein | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Maisonneuve, P, Kurinov, I, Wiechmann, S, Ernst, A, Sicheri, F. | | Deposit date: | 2019-01-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-specific inhibitors of activated Ras GTPases reveal limited Ras dependency of patient-derived cancer organoids.
J.Biol.Chem., 295, 2020
|
|
8JC9
 
 | | Cryo-EM structure of the LH1 complex from thermochromatium tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, DODECANE, ... | | Authors: | Wang, G.-L, Yan, Y.-H, Yu, L.-J. | | Deposit date: | 2023-05-10 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular structure and characterization of the Thermochromatium tepidum light-harvesting 1 photocomplex produced in a foreign host.
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9K92
 
 | | Cryo-EM structure of human signal peptide peptidase like 2A (SPPL2a) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Signal peptide peptidase-like 2A | | Authors: | Zhou, R, Huang, G, Guo, X, Wang, J, Ge, X, Shi, Y. | | Deposit date: | 2024-10-25 | | Release date: | 2025-10-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into human signal peptide peptidase
To Be Published
|
|
7TFQ
 
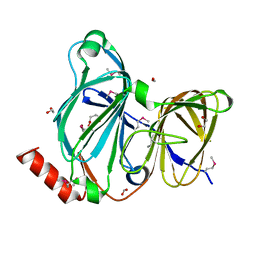 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, FORMIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK Bound to Copper Ion from Yersinia pestis
To Be Published
|
|
8OIG
 
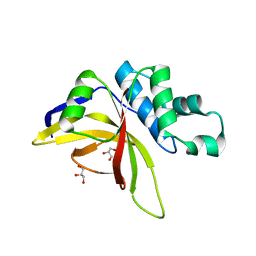 | | Crystal Structure of Staphopain C from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McEwen, A.G, Magoch, M, Napolitano, V, Dubin, G, Wladyka, B. | | Deposit date: | 2023-03-22 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Staphopain C from Staphylococcus aureus.
Molecules, 28, 2023
|
|
8OW3
 
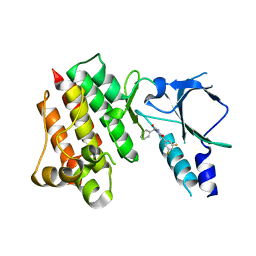 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
7BJI
 
 | | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Centrosomal protein of 135 kDa, ... | | Authors: | Li, Q, Hatzopoulos, G, Iller, O, Vakonakis, I. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190
To Be Published
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
7Q5W
 
 | | The tandem SH2 domains of SYK with a bound TYROBP diphospho-ITAM peptide | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Chen, Z, Bountra, C, von Delft, F, Gileadi, O, Brennan, P.E. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-24 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures.
Structure, 32, 2024
|
|
8H5V
 
 | | Crystal structure of the FleQ domain of Vibrio cholerae FlrA | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, flagellar regulatory protein A | | Authors: | Dasgupta, J, Chakraborty, S. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The N-terminal FleQ domain of the Vibrio cholerae flagellar master regulator FlrA plays pivotal structural roles in stabilizing its active state.
Febs Lett., 597, 2023
|
|
7BRA
 
 | | Bacillus subtilis IRG1 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Bacillus subtilis IRG1, SULFATE ION | | Authors: | Park, H.H, Chun, H.L. | | Deposit date: | 2020-03-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Enzymatic reaction mechanism of cis-aconitate decarboxylase based on the crystal structure of IRG1 from Bacillus subtilis.
Sci Rep, 10, 2020
|
|
6X1Z
 
 | | Mre11 dimer in complex with small molecule modulator PFMJ | | Descriptor: | (5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
8V2H
 
 | |
8V3G
 
 | |
6XQQ
 
 | | Structure of human D462-E4 TCR | | Descriptor: | GLYCEROL, TRAV12-2 alpha chain, TRBV29-1 | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Atypical TRAV1-2 - T cell receptor recognition of the antigen-presenting molecule MR1.
J.Biol.Chem., 295, 2020
|
|
8UKI
 
 | | Crystal structure of 04_A06 Fab | | Descriptor: | 04_A06 Fab Heavy Chain, 04_A06 Fab Light Chain | | Authors: | DeLaitsch, A.T, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2023-10-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Profiling of HIV-1 elite neutralizer cohort reveals a CD4bs bnAb for HIV-1 prevention and therapy.
Nat.Immunol., 2025
|
|
8P9C
 
 | | Crystal structure of p63-p73 heterotetramer (tetramerisation domain) in complex with darpin 1810 F11 | | Descriptor: | 1,2-ETHANEDIOL, Darpin 1810 F11, Tumor protein 63, ... | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DARPins detect the formation of hetero-tetramers of p63 and p73 in epithelial tissues and in squamous cell carcinoma.
Cell Death Dis, 14, 2023
|
|
6WFN
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
8V1V
 
 | |
7SV5
 
 | | Crystal structure of SpaA-SLH/G109A in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
8T68
 
 | | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 in complex with RQ3-111 | | Descriptor: | 6,7-dichloro-N-[(3R)-oxolan-3-yl]-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase KMT5B, S-ADENOSYLMETHIONINE, ... | | Authors: | Zeng, H, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-15 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 in complex with RQ3-111.
To be published
|
|
8X06
 
 | | Cryo-EM structure of the IR/IGF-I complex, conformation 1 | | Descriptor: | Insulin-like growth factor I, Isoform Short of Insulin receptor | | Authors: | Xi, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structure of the IR/IGF-I complex, conformation 1
To Be Published
|
|
6WGX
 
 | | Cocrystal of BRD4(D1) with a selective inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6WJP
 
 | | Crystal structure of Arginine Repressor P115Q mutant from the pathogenic bacterium Corynebacterium pseudotuberculosis bound to arginine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ARGININE, ... | | Authors: | Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, de Morais, M.A.B, Murakami, M.T, Carareto, C.M.A, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A single P115Q mutation modulates specificity in the Corynebacterium pseudotuberculosis arginine repressor.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
