2FQ8
 
 | |
3VH9
 
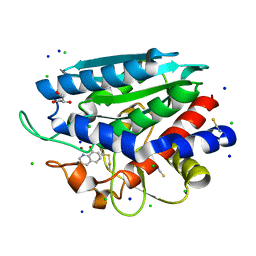 | | Crystal structure of Aeromonas proteolytica aminopeptidase complexed with 8-quinolinol | | Descriptor: | Bacterial leucyl aminopeptidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Hanaya, K, Suetsugu, M, Kobayashi, K, Yamato, I, Aoki, S. | | Deposit date: | 2011-08-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Potent inhibition of dinuclear zinc(II) peptidase, an aminopeptidase from Aeromonas proteolytica, by 8-quinolinol derivatives: inhibitor design based on Zn(2+) fluorophores, kinetic, and X-ray crystallographic study.
J.Biol.Inorg.Chem., 17, 2012
|
|
3B21
 
 | |
4E50
 
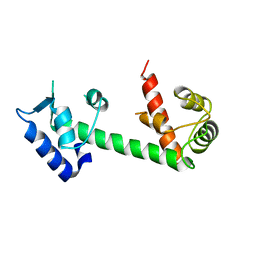 | | Calmodulin and Ng peptide complex | | Descriptor: | Calmodulin, Linker, IQ motif of Neurogranin | | Authors: | Kumar, V, Sivaraman, J. | | Deposit date: | 2012-03-13 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction of unstructured neuron specific substrates neuromodulin and neurogranin with calmodulin
Sci Rep, 3, 2013
|
|
2HW0
 
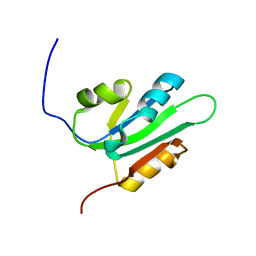 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
1GE8
 
 | |
4Q5H
 
 | |
2DYW
 
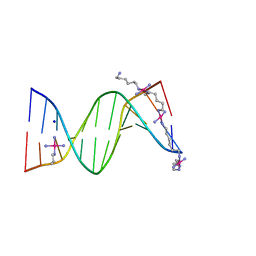 | | A Backbone binding DNA complex | | Descriptor: | (6-AMINOHEXYLAMINE)(TRIAMMINE) PLATINUM(II) COMPLEX, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', SODIUM ION, ... | | Authors: | Komeda, S, Moulaei, T, Woods, K.K, Chikuma, M, Farrell, N.P, Williams, L.D. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A Third Mode of DNA Binding: Phosphate Clamps by a Polynuclear Platinum Complex
J.Am.Chem.Soc., 128, 2006
|
|
5U5R
 
 | | Crystal Structure and X-ray Diffraction Data Collection of Importin-alpha from Mus musculus Complexed with a PMS2 NLS Peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Importin subunit alpha-1, Mismatch repair endonuclease PMS2 | | Authors: | Barros, A.C, Takeda, A.A, Dreyer, T.R, Velazquez-Campoy, A, Kobe, B, Fontes, M.R. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA mismatch repair proteins MLH1 and PMS2 can be imported to the nucleus by a classical nuclear import pathway.
Biochimie, 146, 2018
|
|
1HYH
 
 | |
4R17
 
 | | Ligand-induced aziridine-formation at subunit beta5 of the yeast 20S proteasome | | Descriptor: | (2S,3S)-3-methylaziridine-2-carboxylic acid, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7OR6
 
 | |
7ORD
 
 | |
3LSW
 
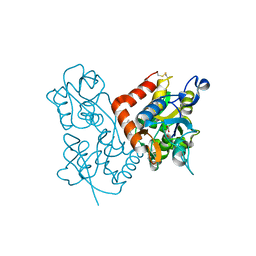 | | Aniracetam bound to the ligand binding domain of GluA3 | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, GLUTAMIC ACID, GluA2 S1S2 domain, ... | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2010-02-13 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
4QR9
 
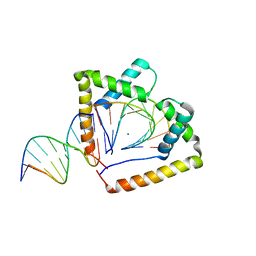 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | Authors: | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XHD
 
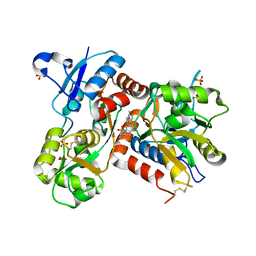 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
4R18
 
 | | Ligand-induced Lys33-Thr1 crosslinking at subunit beta5 of the yeast 20S proteasome | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1HJ7
 
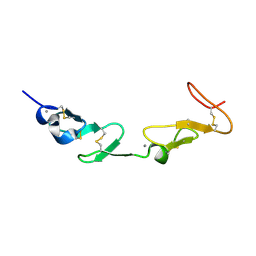 | | NMR study of a pair of LDL receptor Ca2+ binding epidermal growth factor-like domains, 20 structures | | Descriptor: | CALCIUM ION, LDL RECEPTOR | | Authors: | Saha, S, Handford, P.A, Campbell, I.D, Downing, A.K. | | Deposit date: | 2001-01-09 | | Release date: | 2001-07-11 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ldl Receptor Egf-Ab Pair: A Paradigm for the Assembly of Tandem Calcium Binding Egf Domains
Structure, 9, 2001
|
|
7WT6
 
 | |
6PIS
 
 | |
6P9B
 
 | |
7P2V
 
 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a 4-chlorophenyl-spiroindoline capped hydroxamate-based inhibitor, bound to a novel site | | Descriptor: | 5-[[(2S)-2-(4-chlorophenyl)-1'-methyl-spiro[2H-indole-3,4'-piperidine]-1-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, DI(HYDROXYETHYL)ETHER, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
7P2T
 
 | | Tetartohedrally twinned crystal structure of Schistosoma mansoni HDAC8 in complex with a tricyclic thieno[3,2-b]indole capped hydroxamate-based inhibitor, bromine derivative | | Descriptor: | 5-[[(2R)-7-bromanyl-2-phenyl-2,3-dihydrothieno[3,2-b]indol-4-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, CHLORIDE ION, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
7P2U
 
 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a 3-chlorophenyl-spiroindoline capped hydroxamate-based inhibitor, bound to a novel site | | Descriptor: | 5-[[(2R)-2-(3-chlorophenyl)-1'-methyl-spiro[2H-indole-3,4'-piperidine]-1-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, CHLORIDE ION, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
7P2S
 
 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a tricyclic thieno[3,2-b]indole capped hydroxamate-based inhibitor, chlorine derivative | | Descriptor: | 5-[[(2R)-7-fluoranyl-2-phenyl-2,3-dihydrothieno[3,2-b]indol-4-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
