9EV8
 
 | |
7NUP
 
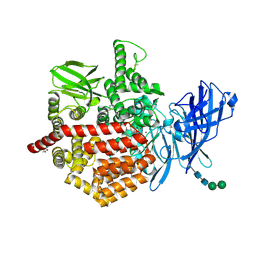 | | Endoplasmic reticulum aminopeptidase 2 complexed with a mixed hydroxamic and sulfonyl ligand | | Descriptor: | (3~{S})-4-(4-hydroxyphenyl)-~{N}-oxidanyl-3-[5-[[(5-phenylthiophen-2-yl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]butanamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2021-03-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endoplasmic reticulum aminopeptidase 2 complexed with a mixed hydroxamic and sulfonyl ligand
To Be Published
|
|
7NSK
 
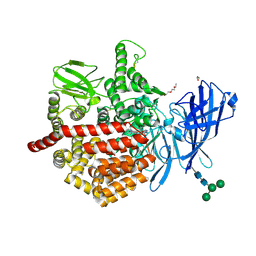 | | Endoplasmic reticulum aminopeptidase 2 complexed with a hydroxamic ligand | | Descriptor: | (2~{S})-3-(4-hydroxyphenyl)-~{N}-oxidanyl-2-[4-[[(5-pyridin-2-ylthiophen-2-yl)sulfonylamino]methyl]-1,2,3-triazol-1-yl]propanamide, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Mpakali, A, Giastas, P, Stratikos, E. | | Deposit date: | 2021-03-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Endoplasmic reticulum aminopeptidase 2 in complex with a phosphinic ligand
To Be Published
|
|
9ECR
 
 | |
9ECQ
 
 | |
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4XDK
 
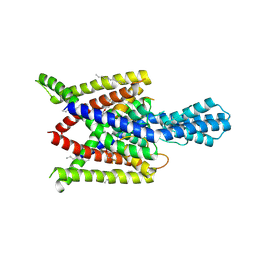 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) in complex with norfluoxetine | | Descriptor: | (3R)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, (3S)-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Mackenzie, A, Mukhopadhyay, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-19 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac.
Science, 347, 2015
|
|
6NRP
 
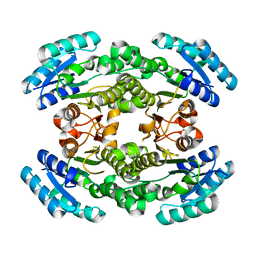 | | Putative short-chain dehydrogenase/reductase (SDR) from Acinetobacter baumannii | | Descriptor: | 3-oxoacyl-ACP reductase FabG | | Authors: | Cross, E.M, Smith, K.M, Shaw, K.I, Aragao, D, Forwood, J.K. | | Deposit date: | 2019-01-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Acinetobacter baumannii fatty acid synthesis 3-oxoacyl-ACP reductases.
Sci Rep, 11, 2021
|
|
8V36
 
 | |
4XGH
 
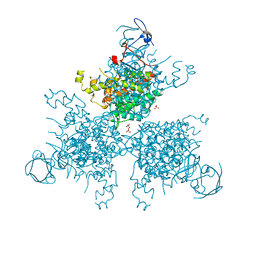 | |
8V35
 
 | | Crystal structure of HpsN from Cupriavidus pinatubonensis | | Descriptor: | 1,2-ETHANEDIOL, Sulfopropanediol 3-dehydrogenase, ZINC ION | | Authors: | Lee, M. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and kinetic insights into the stereospecific oxidation of R -2,3-dihydroxypropanesulfonate by DHPS-3-dehydrogenase from Cupriavidus pinatubonensis.
Chem Sci, 15, 2024
|
|
4XBM
 
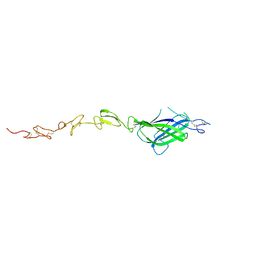 | | X-ray crystal structure of Notch ligand Delta-like 1 | | Descriptor: | Delta-like protein 1, alpha-L-fucopyranose | | Authors: | Kershaw, N.J, Burgess, A.W, Church, N.L, Luo, C.S, Adam, T.E. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Notch ligand delta-like1: X-ray crystal structure and binding affinity.
Biochem.J., 468, 2015
|
|
6WKF
 
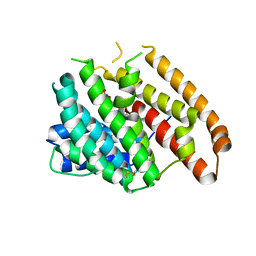 | | Crystal structure of pentalenene synthase mutant F76Y complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
4XP4
 
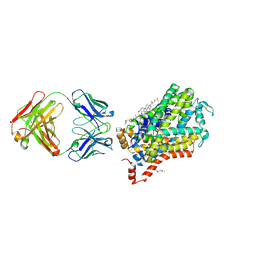 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7PTI
 
 | |
6WXP
 
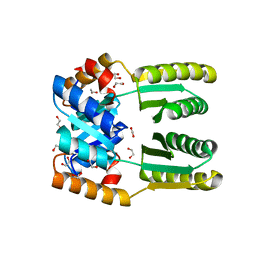 | | De novo TIM barrel-ferredoxin fold fusion homodimer with 4-glutamate centre TFD-EE | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TFD-EE | | Authors: | Caldwell, S.J, Zeymer, C, Haydon, I.C, Huang, P, Hilvert, D, Baker, D. | | Deposit date: | 2020-05-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XKX
 
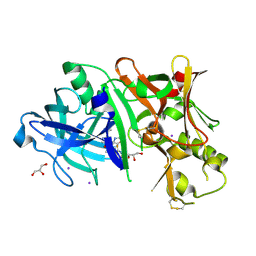 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-azaxanthene inhibitor 28 | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-(2-fluoropyridin-4-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious beta-secretase inhibitors for the potential treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6WN0
 
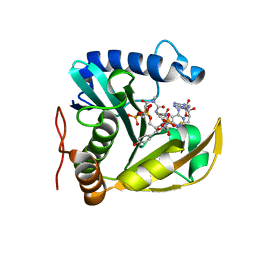 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
4XR8
 
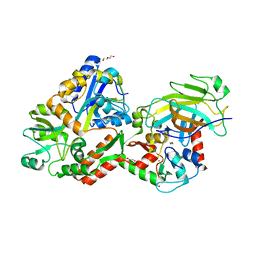 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
6WKJ
 
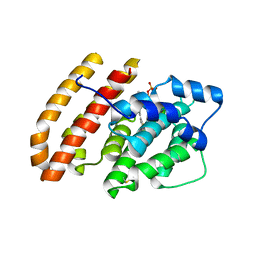 | | Crystal structure of pentalenene synthase mutant F76H complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
6WKH
 
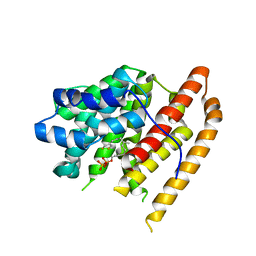 | | Crystal structure of pentalenene synthase mutant F76W complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
5FAA
 
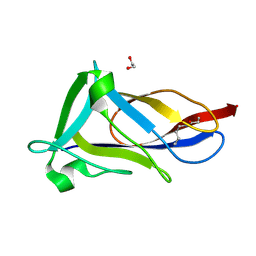 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
8EYU
 
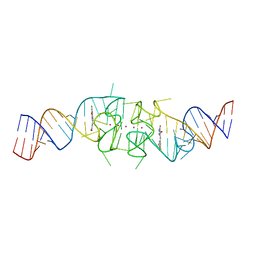 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
7T8K
 
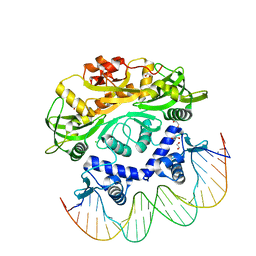 | | BrxR from Acinetobacter BREX type I phage restriction system bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, BrxR, CHLORIDE ION, ... | | Authors: | Doyle, L, Kaiser, B, Stoddard, B. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and characterization of the WYL BrxR protein and its gene as separable regulatory elements of a BREX phage restriction system.
Nucleic Acids Res., 50, 2022
|
|
