6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
6JMD
 
 | | Crystal structure of human DHODH in complex with inhibitor 1223 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-[3-(hydroxymethyl)phenyl]phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
5AR0
 
 | | HSP72 with adenosine-derived inhibitor | | Descriptor: | (2R,3R,4S,5R)-2-(6-amino-8-((quinolin-7-ylmethyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Cheeseman, M.D, Westwood, I.M, Barbeau, O, Rowlands, M.G, Jones, A.M, Jeganathan, F, Burke, R, Dobson, S.E, Workman, P, Collins, I, van Montfort, R.L.M, Jones, K. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of Hsp70.
J.Med.Chem., 59, 2016
|
|
6HZU
 
 | | HUMAN JAK1 IN COMPLEX WITH LASW1393 | | Descriptor: | 2-[4-[8-oxidanylidene-2-[(~{E})-(2-oxidanylidenepyridin-3-ylidene)amino]-7~{H}-purin-9-yl]cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Lozoya, E, Segarra, V, Bach, J, Jestel, A, Lammens, A, Blaesse, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of 2-Imidazopyridine and 2-Aminopyridone Purinones as Potent Pan-Janus Kinase (JAK) Inhibitors for the Inhaled Treatment of Respiratory Diseases.
J.Med.Chem., 62, 2019
|
|
6I6F
 
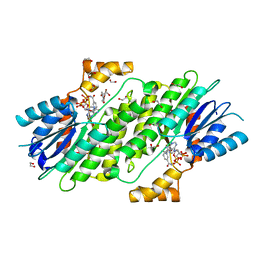 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6V
 
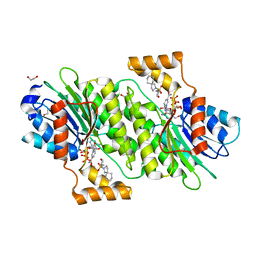 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(3~{R})-oxan-3-yl]methylsulfonyl]-2-azaspiro[4.5]decane, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
1QXD
 
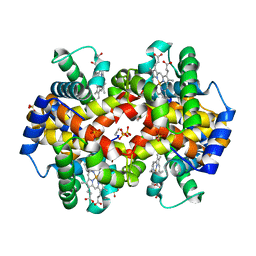 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
1QXE
 
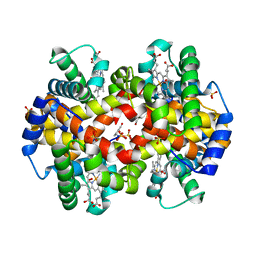 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | 5-HYDROXYMETHYL-FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
6RIH
 
 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ2
 
 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
7PDR
 
 | | Crystal structure of Lymnaea stagnalis Acetylcholine-binding protein (Ls-AChBP) Q55R/M114V double mutant complexed with Dichloromezotiaz | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]-1-[(2-chloranyl-1,3-thiazol-5-yl)methyl]-9-methyl-pyrido[1,2-a]pyrimidine-2,4-dione, Acetylcholine-binding protein | | Authors: | Montgomery, M.G. | | Deposit date: | 2021-08-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Biology-Guided Design, Synthesis, and Biological Evaluation of Novel Insect Nicotinic Acetylcholine Receptor Orthosteric Modulators.
J.Med.Chem., 65, 2022
|
|
7PDB
 
 | |
7PE5
 
 | |
7PE6
 
 | |
7Q5H
 
 | | Keap1 compound complex | | Descriptor: | (3S,5S,8R)-8-[[(2S)-1-ethanoylpyrrolidin-2-yl]carbonylamino]-N,N-dimethyl-3,7,11-tris(oxidanylidene)-10-oxa-3$l^{4}-thia-6-azabicyclo[10.4.0]hexadeca-1(16),12,14-triene-5-carboxamide, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | Authors: | Johansson, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
1NZ7
 
 | | POTENT, SELECTIVE INHIBITORS OF PROTEIN TYROSINE PHOSPHATASE 1B USING A SECOND PHOSPHOTYROSINE BINDING SITE, complexed with compound 19. | | Descriptor: | 2-[(4-{2-ACETYLAMINO-2-[4-(1-CARBOXY-3-METHYLSULFANYL-PROPYLCARBAMOYL)-BUTYLCARBAMOYL]-ETHYL}-2-ETHYL-PHENYL)-OXALYL-AM INO]-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Xin, Z, Oost, T.K, Abad-Zapatero, C, Hajduk, P.J, Pei, Z, Szczepankiewicz, B.G, Hutchins, C.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Trevillyan, J.M, Jirousek, M.R, Liu, G. | | Deposit date: | 2003-02-16 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent, Selective Inhibitors of Protein Tyrosine Phosphatase 1B
BIOORG.MED.CHEM.LETT., 13, 2003
|
|
3M3F
 
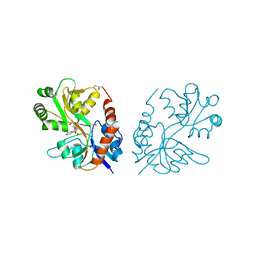 | | PEPA bound to the ligand binding domain of GluA3 (flop form) | | Descriptor: | 2-[2,6-difluoro-4-({2-[(phenylsulfonyl)amino]ethyl}sulfanyl)phenoxy]acetamide, GLUTAMIC ACID, Glutamate receptor 3, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of flop selectivity and subsite recognition for an AMPA receptor allosteric modulator: structures of GluA2 and GluA3 in complexes with PEPA.
Biochemistry, 49, 2010
|
|
7Q5O
 
 | | Bromodomain-containing 2 BD2 in complex with the inhibitor CRCM5484 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[11-ethanoyl-4-(furan-2-ylmethyl)-3-oxidanylidene-8-thia-4,6,11-triazatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),5-trien-5-yl]sulfanyl]-~{N}-(2-methylpyridin-3-yl)ethanamide, Bromodomain-containing protein 2 | | Authors: | Carrasco, K, Betzi, S, Morelli, X. | | Deposit date: | 2021-11-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | CRCM5484: A BET-BDII Selective Compound with Differential Anti-leukemic Drug Modulation
J.Med.Chem., 65, 2022
|
|
4V2O
 
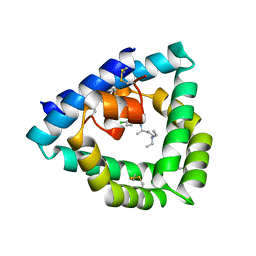 | | Structure of saposin B in complex with chloroquine | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, SAPOSIN-B | | Authors: | Zubieta, C, Lai, X, Doyle, R.P. | | Deposit date: | 2014-10-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Lysosomal Protein Saposin B Binds Chloroquine.
Chemmedchem, 11, 2016
|
|
7PTF
 
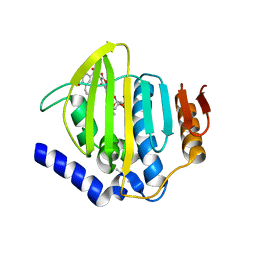 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PTG
 
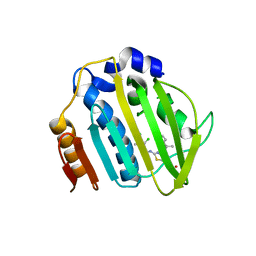 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQI
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQM
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7W47
 
 | | Crystal structure of the gastric proton pump complexed with tegoprazan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Potassium-transporting ATPase alpha chain 1, ... | | Authors: | Abe, K, Tanaka, S, Morita, M, Yamagishi, T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump.
J.Med.Chem., 65, 2022
|
|
