4UXD
 
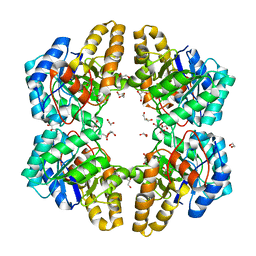 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
6NOD
 
 | | Crystal structure of C. elegans PUF-8 in complex with RNA | | Descriptor: | PUF (Pumilio/FBF) domain-containing, RNA (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3') | | Authors: | Wang, Y, McCann, K.L, Qiu, C, Hall, T.M.T. | | Deposit date: | 2019-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Engineering a conserved RNA regulatory protein repurposes its biological function in vivo .
Elife, 8, 2019
|
|
6NPN
 
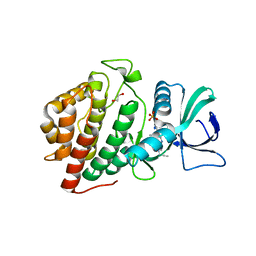 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor | | Descriptor: | (7R)-7-methyl-2-[(3-oxo-2,3-dihydro-1H-indazol-6-yl)amino]-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, dos Reis, C.V, Takarada, J.E, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor
To Be Published
|
|
4UY4
 
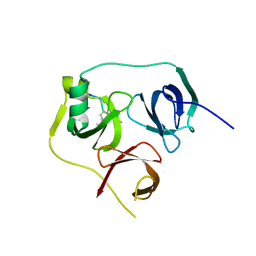 | | 1.86 A structure of human Spindlin-4 protein in complex with histone H3K4me3 peptide | | Descriptor: | GLYCEROL, HISTONE H3K4ME3, SPINDLIN-4 | | Authors: | Talon, R, Gileadi, C, Johansson, C, Burgess-Brown, N, Shrestha, L, von Delft, F, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | 1.86 A Structure of Human Spindlin-4 Protein in Complex with Histone H3K4Me3 Peptide
To be Published
|
|
6SEB
 
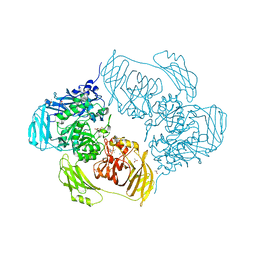 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
4V19
 
 | | Structure of the large subunit of the mammalian mitoribosome, part 1 of 2 | | Descriptor: | MAGNESIUM ION, MITORIBOSOMAL 16S RRNA, MITORIBOSOMAL CP TRNA, ... | | Authors: | Greber, B.J, Boehringer, D, Leibundgut, M, Bieri, P, Leitner, A, Schmitz, N, Aebersold, R, Ban, N. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Complete Structure of the Large Subunit of the Mammalian Mitochondrial Ribosome
Nature, 515, 2014
|
|
6NR3
 
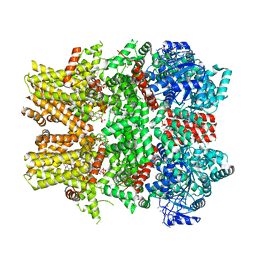 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NS7
 
 | |
6NSE
 
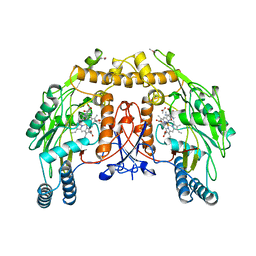 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, CANAVANINE COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
6NMG
 
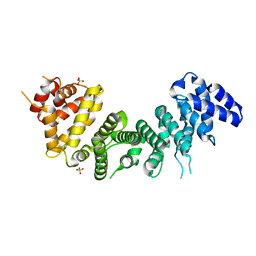 | | Crystal Structure of Rat Ric-8A G alpha binding domain | | Descriptor: | Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans), SULFATE ION | | Authors: | Zeng, B, Mou, T.C, Sprang, S.R. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Function, and Dynamics of the G alpha Binding Domain of Ric-8A.
Structure, 27, 2019
|
|
6NMQ
 
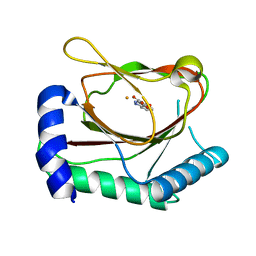 | | Hypoxia-Inducible Factor (HIF) Prolyl Hydroxylase 2 (PHD2) in Complex with the Carboxamide Analog JNJ43058171 | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-(4-oxo-1,4-dihydrocinnoline-3-carbonyl)glycine | | Authors: | Bembenek, S.D, Mirzadegan, T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Beyond Traditional Structure-Based Drug Design: The Role of Iron Complexation, Strain, and Water in the Binding of Inhibitors for Hypoxia-Inducible Factor Prolyl Hydroxylase 2.
Acs Omega, 4, 2019
|
|
6NSP
 
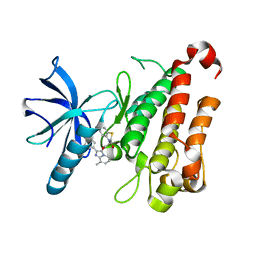 | | TRK-A IN COMPLEX WITH LIGAND 9 | | Descriptor: | High affinity nerve growth factor receptor, N-(8-methyl-2-phenylimidazo[1,2-a]pyridin-3-yl)-2-(3-oxo-2,3-dihydro-4H-1,4-benzothiazin-4-yl)acetamide | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Deciphering the Allosteric Binding Mechanism of the Human Tropomyosin Receptor Kinase A ( hTrkA) Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
6NN8
 
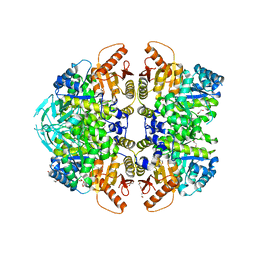 | | The structure of human liver pyruvate kinase, hLPYK-S531E | | Descriptor: | 1,2-ETHANEDIOL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6SN6
 
 | |
6NPP
 
 | |
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6NUS
 
 | |
6NV2
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
4WE0
 
 | | JC Polyomavirus VP1 five-fold pore mutant P223M | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of a Pore in the Capsid of JC Polyomavirus Reduces Infectivity and Prevents Exposure of the Minor Capsid Proteins.
J.Virol., 89, 2015
|
|
6NQB
 
 | |
4UYT
 
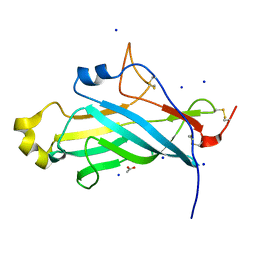 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, FLOCCULATION PROTEIN FLO11, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4UQX
 
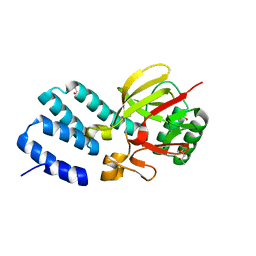 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
1E6N
 
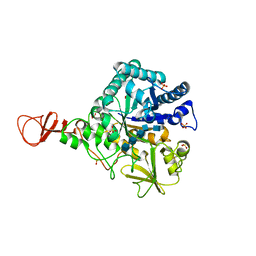 | | Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE B, GLYCEROL, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6NBY
 
 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
