2GHZ
 
 | |
2GI0
 
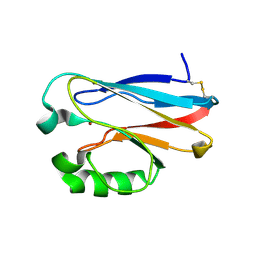 | |
2GI3
 
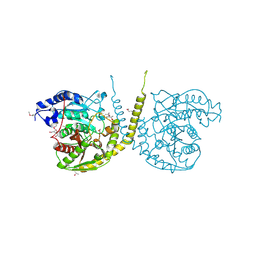 | |
2GI4
 
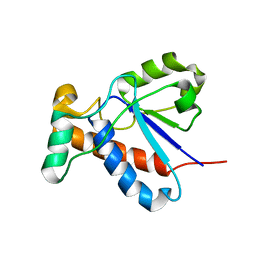 | |
2GI7
 
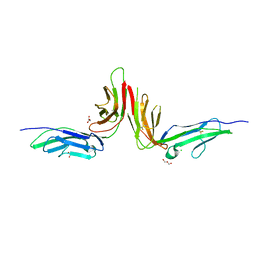 | | Crystal structure of human platelet Glycoprotein VI (GPVI) | | Descriptor: | CHLORIDE ION, GLYCEROL, GPVI protein, ... | | Authors: | Horii, K, Kahn, M.L, Herr, A.B. | | Deposit date: | 2006-03-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI.
Blood, 108, 2006
|
|
2GI9
 
 | |
2GIA
 
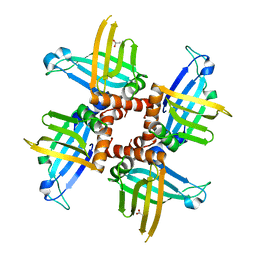 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | ACETIC ACID, mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
2GIB
 
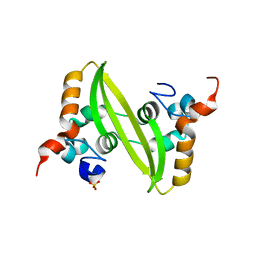 | | Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain | | Descriptor: | Nucleocapsid protein, SULFATE ION | | Authors: | Yu, I.M, Oldham, M.L, Zhang, J, Chen, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae.
J.Biol.Chem., 281, 2006
|
|
2GIC
 
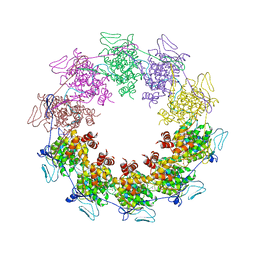 | | Crystal Structure of a vesicular stomatitis virus nucleocapsid-RNA complex | | Descriptor: | 45-MER, Nucleocapsid protein, URANYL (VI) ION | | Authors: | Green, T.J, Zhang, X, Wertz, G.W, Luo, M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structure of the vesicular stomatitis virus nucleoprotein-RNA complex unveils how the RNA is sequestered
Science, 313, 2006
|
|
2GID
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
2GIE
 
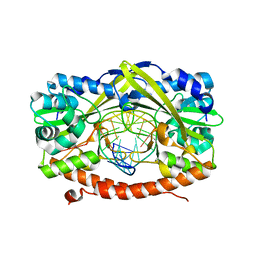 | | HincII bound to cognate DNA GTTAAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*G)-3', SODIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GIF
 
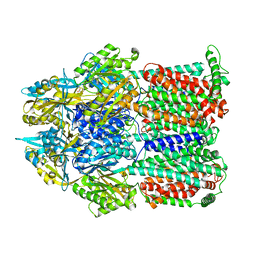 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
2GIG
 
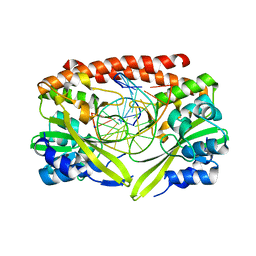 | | Alteration of sequence specificity of the type II restriction endonuclease HINCII through an indirect readout mechanism | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', SODIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GIH
 
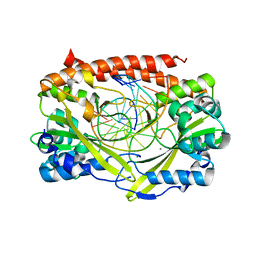 | | Q138F HincII bound to cognate DNA GTCGAC and Ca2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', CALCIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GII
 
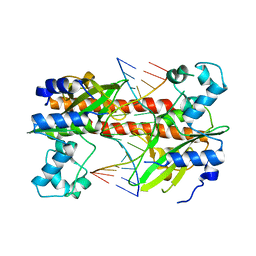 | | Q138F HincII bound to cognate DNA GTTAAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*GP*C)-3', Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GIJ
 
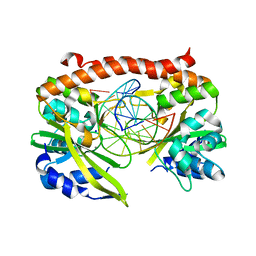 | | Q138F HincII bound to cognate DNA GTTAAC and Ca2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*GP*C)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Horton, N.C, Joshi, H.K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
2GIL
 
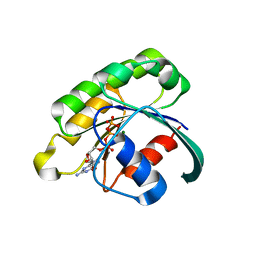 | | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-6A | | Authors: | Bergbrede, T, Pylypenko, O, Rak, A, Alexandrov, K. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 resolution
J.STRUCT.BIOL., 152, 2005
|
|
2GIM
 
 | |
2GIN
 
 | |
2GIO
 
 | |
2GIP
 
 | |
2GIQ
 
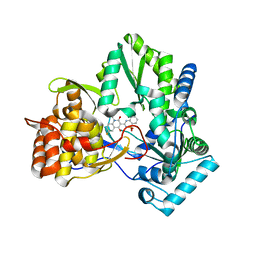 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-2 inhibitor | | Descriptor: | 1-(2-CYCLOPROPYLETHYL)-3-(1,1-DIOXIDO-2H-1,2,4-BENZOTHIADIAZIN-3-YL)-6-FLUORO-4-HYDROXYQUINOLIN-2(1H)-ONE, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GIR
 
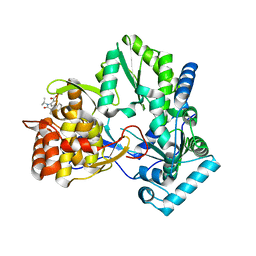 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-1 inhibitor | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
2GIS
 
 | |
2GIT
 
 | |
