8DJK
 
 | | HMGCR-UBIAD1 Complex State 2 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
8WMP
 
 | | Crystal Structure of Mutant HisB from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tiwari, S, Mohini, M, Ahmad, M, Pal, R.K, Biswal, B.K. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mutant HisB from Mycobacterium tuberculosis
To Be Published
|
|
8B0B
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8B0C
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB158 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-3,4,6-tris(oxidanyl)-2-[2-[2,2,2-tris(fluoranyl)ethanoylamino]ethoxy]cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase 50 kDa subunit, ... | | Authors: | Armstrong, Z, Davies, G. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
5MTQ
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT511 | | Descriptor: | 2-[4-[(4-cyclohexyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
8R66
 
 | | Crystal structure of ThsA Macro domain in complex with signaling molecule | | Descriptor: | (2~{S})-3-[1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]imidazol-4-yl]-2-azanyl-propanoic acid, SULFATE ION, Thoeris protein ThsA | | Authors: | Tamulaitiene, G, Sabonis, D. | | Deposit date: | 2023-11-21 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | TIR domains produce histidine-ADPR as an immune signal in bacteria.
Nature, 642, 2025
|
|
8FH9
 
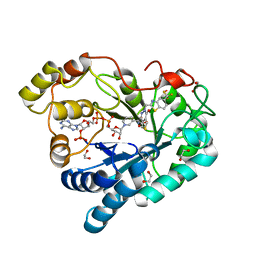 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | Descriptor: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
6NTK
 
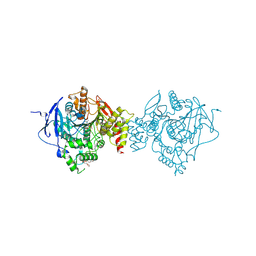 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-232 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
6YR3
 
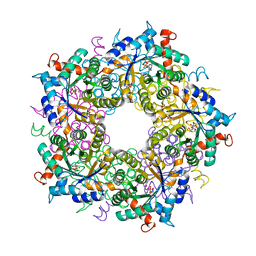 | |
5JOV
 
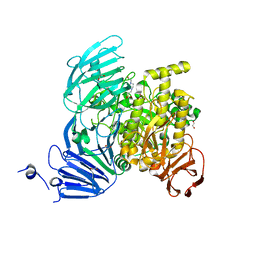 | | Bacteroides ovatus Xyloglucan PUL GH31 with bound 5FIdoF | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-fluoro-alpha-L-idopyranose, ... | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6O0P
 
 | | crystal structure of BCL-2 G101A mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
7PGN
 
 | | HHP-C in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Griffiths, S.C, Schwab, R.A, El Omari, K, Bishop, B, Iverson, E.J, Malinuskas, T, Dubey, R, Qian, M, Covey, D.F, Gilbert, R.J.C, Rohatgi, R, Siebold, C. | | Deposit date: | 2021-08-14 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hedgehog-Interacting Protein is a multimodal antagonist of Hedgehog signalling.
Nat Commun, 12, 2021
|
|
8SAE
 
 | |
6EPV
 
 | | The ATAD2 bromodomain in complex with compound 5 | | Descriptor: | (2~{R})-2-azanyl-~{N}-(4-oxidanylidene-6,7-dihydro-5~{H}-1,3-benzothiazol-2-yl)propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
8OOW
 
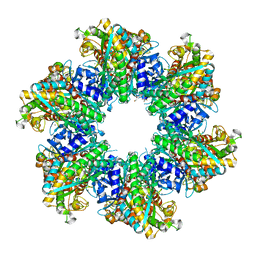 | | Glutamine synthetase from Methermicoccus shengliensis at a resolution of 2.64 A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glutamine synthetase, ... | | Authors: | Mueller, M.-C, Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations.
Commun Biol, 7, 2024
|
|
6O0K
 
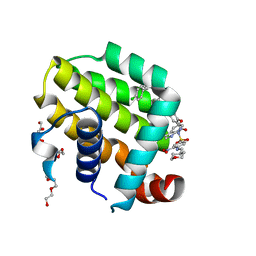 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6QNC
 
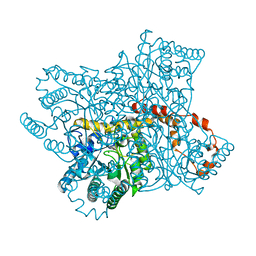 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 0.1 s timepoint | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | Deposit date: | 2019-02-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6HEH
 
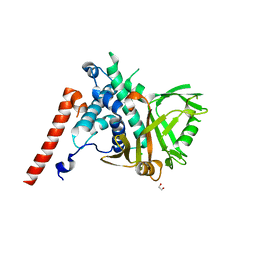 | | Structure of the catalytic domain of USP28 (insertion deleted) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28,Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
8D7N
 
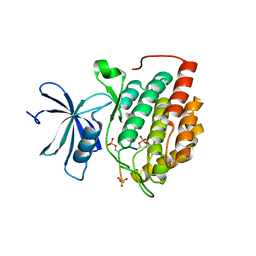 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
6O66
 
 | |
6O9U
 
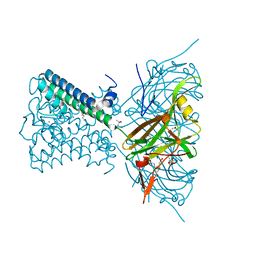 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6ABH
 
 | |
7PH3
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
6OBA
 
 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
