8CCV
 
 | | E. coli NfsB mutant T41LN71S with nicotinate | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Day, M.A, White, S.A, Hyde, E.I, Searle, P.F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
8CJ0
 
 | | E. coli NfsB-T41Q/N71S/F124T/M127V mutant bound to nicotinate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NICOTINIC ACID, ... | | Authors: | White, S.A, Hyde, E.I, Day, M.A. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and Dynamics of Three Escherichia coli NfsB Nitro-Reductase Mutants Selected for Enhanced Activity with the Cancer Prodrug CB1954.
Int J Mol Sci, 24, 2023
|
|
2WSE
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure determination and improved model of plant photosystem I
J. Biol. Chem., 285, 2010
|
|
6KOL
 
 | | Crystal structure of auracyanin from photosynthetic bacterium Roseiflexus castenholzii | | Descriptor: | Blue (Type 1) copper domain protein, CHLORIDE ION, COPPER (II) ION | | Authors: | Wang, C, Zhang, C.Y, Min, Z.Z, Xin, Y.Y, Xu, X.L. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structural basis underlying the electron transfer features of a blue copper protein auracyanin from the photosynthetic bacterium Roseiflexus castenholzii.
Photosyn. Res., 143, 2020
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
8C78
 
 | | Crystal structure of human BCL6 BTB domain in complex with compound CCT374705 | | Descriptor: | (2~{S})-10-[(3-chloranyl-2-fluoranyl-pyridin-4-yl)amino]-2-cyclopropyl-3,3-bis(fluoranyl)-7-methyl-2,4-dihydro-1~{H}-[1,4]oxazepino[2,3-c]quinolin-6-one, 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, ... | | Authors: | Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an In Vivo Chemical Probe for BCL6 Inhibition by Optimization of Tricyclic Quinolinones.
J.Med.Chem., 66, 2023
|
|
3O3I
 
 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OH at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*U)-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O42
 
 | |
6QZC
 
 | | HLA-DR1 with the QAR Peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
6KSW
 
 | | Cryo-EM structure of the human concentrative nucleoside transporter CNT3 | | Descriptor: | Solute carrier family 28 member 3 | | Authors: | Zhou, Y.X, Liao, L.H, Li, J.L, Xiao, Q.J, Sun, L.F, Deng, D. | | Deposit date: | 2019-08-26 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the human concentrative nucleoside transporter CNT3.
Plos Biol., 18, 2020
|
|
6KVZ
 
 | |
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
6LD8
 
 | |
3NY8
 
 | | Crystal structure of the human beta2 adrenergic receptor in complex with the inverse agonist ICI 118,551 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, Beta-2 adrenergic receptor, ... | | Authors: | Wacker, D, Fenalti, G, Brown, M.A, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
2XQU
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
3NY9
 
 | | Crystal structure of the human beta2 adrenergic receptor in complex with a novel inverse agonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Fenalti, G, Wacker, D, Brown, M.A, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
3NYJ
 
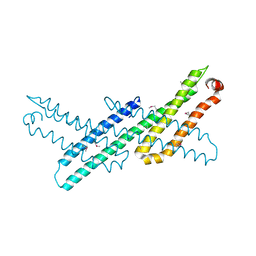 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
8C4O
 
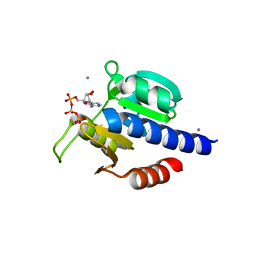 | | CdaA-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Diadenylate cyclase, ... | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
8C4R
 
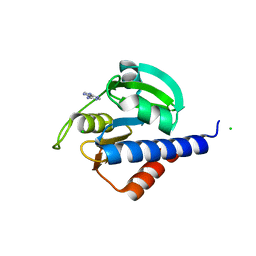 | | CdaA-adenine complex | | Descriptor: | ADENINE, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
8C4J
 
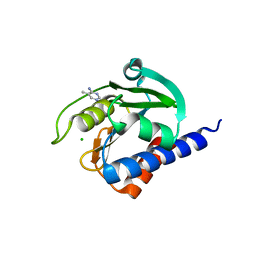 | | CdaA-compound 4 complex | | Descriptor: | 5-methylpyrimidin-4-amine, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
8C4Q
 
 | | CdaA-Apo | | Descriptor: | CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
3O2D
 
 | | Crystal structure of HIV-1 primary receptor CD4 in complex with a potent antiviral antibody | | Descriptor: | T-cell surface glycoprotein CD4, ibalizumab heavy chain, ibalizumab light chain | | Authors: | Freeman, M.M, Seaman, M.S, Rits-Volloch, S, Hong, X, Ho, D.D, Chen, B. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of HIV-1 Primary Receptor CD4 in Complex with a Potent Antiviral Antibody.
Structure, 18, 2010
|
|
3O35
 
 | |
8C4M
 
 | | CdaA-Adenosine complex | | Descriptor: | ADENOSINE, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
6R2D
 
 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD after soaking with succinylphosphonate phosphonoethyl ester, followed by temperature increase | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-[ethoxy(oxidanyl)phosphoryl]-4-oxidanyl-butanoic acid, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2019-03-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J.Struct.Biol., 208, 2019
|
|
