1SAX
 
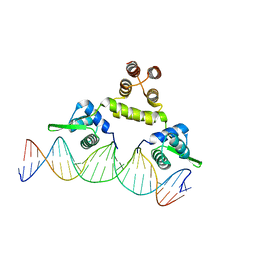 | | Three-dimensional structure of s.aureus methicillin-resistance regulating transcriptional repressor meci in complex with 25-bp ds-DNA | | Descriptor: | 5'-d(CAAAATTACAACTGTAATATCGGAG)-3', 5'-d(GCTCCGATATTACAGTTGTAATTTT)-3', Methicillin resistance regulatory protein mecI, ... | | Authors: | Garcia-Castellanos, R, Mallorqui-Fernandez, G, Marrero, A, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator
J.Biol.Chem., 279, 2004
|
|
1UNO
 
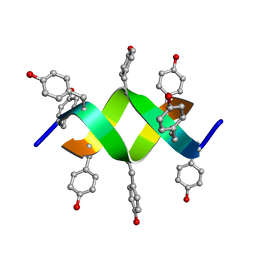 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1J7X
 
 | |
2IWD
 
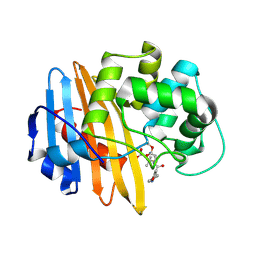 | | Oxacilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Methicillin resistance mecR1 protein | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
2IWB
 
 | | MecR1 unbound extracellular antibiotic-sensor domain. | | Descriptor: | GLYCEROL, METHICILLIN RESISTANCE MECR1 PROTEIN, NICKEL (II) ION, ... | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
1K9V
 
 | | Structural evidence for ammonia tunelling across the (beta-alpha)8-barrel of the imidazole glycerol phosphate synthase bienzyme complex | | Descriptor: | ACETIC ACID, Amidotransferase hisH | | Authors: | Douangamath, A, Walker, M, Beismann-Driemeyer, S, Vega-Fernandez, M.C, Sterner, R, Wilmanns, M. | | Deposit date: | 2001-10-31 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
Structure, 10, 2002
|
|
2IWC
 
 | | Benzylpenicilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | METHICILLIN RESISTANCE MECR1 PROTEIN, OPEN FORM - PENICILLIN G | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
2JML
 
 | | Solution structure of the N-terminal domain of CarA repressor | | Descriptor: | DNA BINDING DOMAIN/TRANSCRIPTIONAL REGULATOR | | Authors: | Jimenez, M, Padmanabhan, S, Gonzalez, C, Perez-Marin, M.C, Elias-Arnanz, M, Murillo, F.J, Rico, M. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor.
Mol.Microbiol., 63, 2007
|
|
1N23
 
 | | (+)-Bornyl diphosphate synthase: Complex with Mg, pyrophosphate, and (1R,4S)-2-azabornane | | Descriptor: | (+)-bornyl diphosphate synthase, (1R,4S)-2-AZABORNANE, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1YDP
 
 | | 1.9A crystal structure of HLA-G | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Clements, C.S, Kjer-nielsen, L, Kostenko, L, Hoare, H.L, Dunstone, M.A, Moses, E, Freed, K, Brooks, A.G, Rossjohn, J, Mccluskey, J. | | Deposit date: | 2004-12-25 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
1MN7
 
 | | NDP kinase mutant (H122G;N119S;F64W) in complex with aBAZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXY-THYMIDINE-5'-ALPHA BORANO TRIPHOSPHATE, MAGNESIUM ION, NDP kinase | | Authors: | gallois-montbrun, s, schneider, b, chen, y, giacomoni-fernandes, v, mulard, l, morera, s, janin, j, deville-bonne, d, veron, m. | | Deposit date: | 2002-09-05 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation
J.BIOL.CHEM., 277, 2002
|
|
1YRR
 
 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
1OKR
 
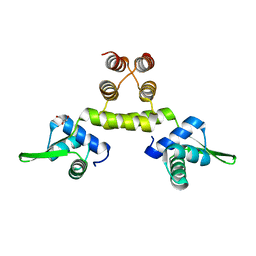 | | Three-dimensional structure of S.aureus methicillin-resistance regulating transcriptional repressor MecI. | | Descriptor: | CHLORIDE ION, GLYCEROL, METHICILLIN RESISTANCE REGULATORY PROTEIN MECI | | Authors: | Garcia-Castellanos, R, Marrero, A, Mallorqui-Fernandez, G, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2003-07-28 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Meci: Molecular Basis for Transcriptional Regulation of Staphylococcal Methicillin Resistance
J.Biol.Chem., 278, 2003
|
|
4CVM
 
 | |
4CVK
 
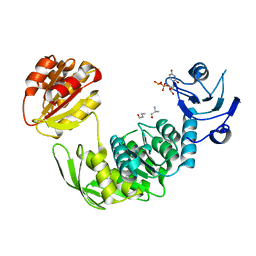 | | PaMurF in complex with UDP-MurNAc-tripeptide (mDAP) | | Descriptor: | ALA-FGA-API, N-acetyl-alpha-muramic acid, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase, ... | | Authors: | Ruane, K.M, Majce, V. | | Deposit date: | 2014-03-28 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Pamurf in Complex with Udp-Murnac-Tripeptide (Mdap)
To be Published
|
|
7V93
 
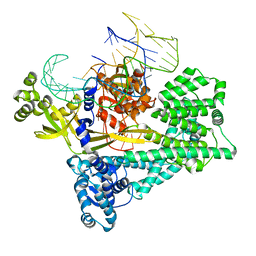 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | Descriptor: | cas12c2, sgRNA | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
1YRG
 
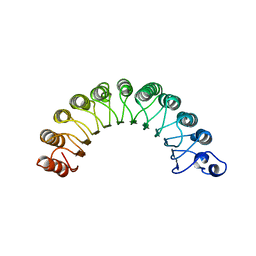 | | THE CRYSTAL STRUCTURE OF RNA1P: A NEW FOLD FOR A GTPASE-ACTIVATING PROTEIN | | Descriptor: | GTPASE-ACTIVATING PROTEIN RNA1_SCHPO | | Authors: | Hillig, R.C, Renault, L, Vetter, I.R, Drell, T, Wittinghofer, A, Becker, J. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The crystal structure of rna1p: a new fold for a GTPase-activating protein.
Mol.Cell, 3, 1999
|
|
7AOE
 
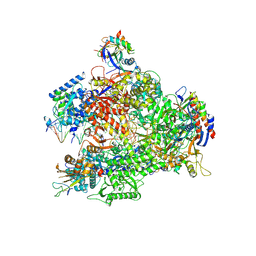 | | Schizosaccharomyces pombe RNA polymerase I (elongation complex) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7S7B
 
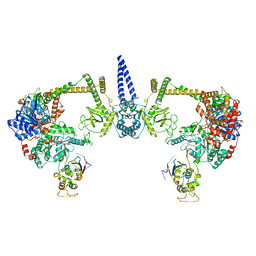 | |
7S7C
 
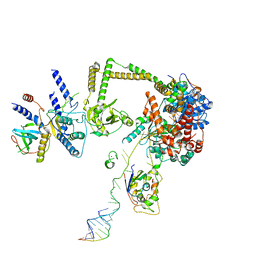 | |
7NVY
 
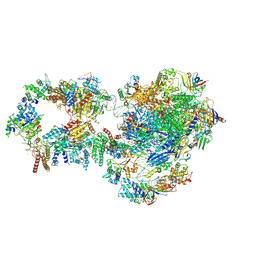 | | RNA polymerase II pre-initiation complex with closed promoter DNA in proximal position | | Descriptor: | CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
205D
 
 | |
1ZSE
 
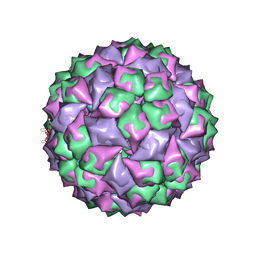 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2K7E
 
 | | NMR structure of the human tRNALys3 bound to the HIV genome Loop I | | Descriptor: | RNA (5'-R(*CP*UP*(SUR)P*UP*UP*AP*AP*(PSU)P*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*GP*GP*UP*GP*UP*AP*AP*AP*AP*G)-3') | | Authors: | Bilbille, Y, Vendeix, F.P.A, Guenther, R, Malkiewicz, A, Ariza, X, Vilarrasa, J, Agris, P. | | Deposit date: | 2008-08-09 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs.
Nucleic Acids Res., 37, 2009
|
|
2JPP
 
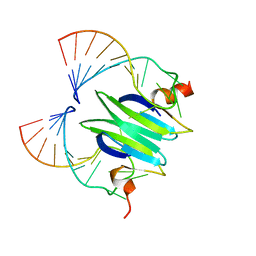 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
