9AXH
 
 | | Crystal structure of KSR1/MEK1 complex heterotetramer with NST-628 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXF
 
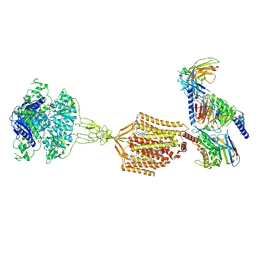 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AXY
 
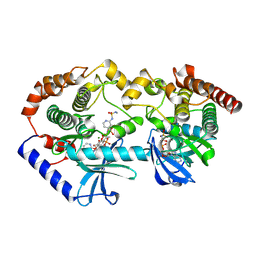 | | Crystal structure of BRAF/MEK complex with NST-628 and inactive RAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXM
 
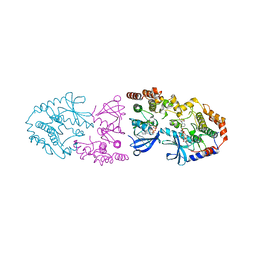 | | Crystal structure of ARAF/MEK1 complex with NST-628 and a RAF dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXC
 
 | | Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXX
 
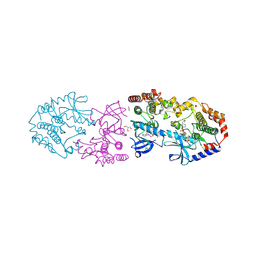 | | Crystal structure of BRAF/MEK1 complex with NST-628 and an active RAF dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
9AXA
 
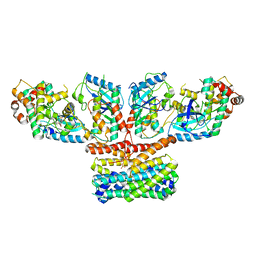 | | CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628 | | Descriptor: | 14-3-3 protein sigma, Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, ... | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
8S8C
 
 | | Structure of Kras in complex with inhibitor MK-1084 | | Descriptor: | (5aSa,17aRa)- 20-Chloro-2-[(2S,5R)-2,5-dimethyl-4-(prop-2-enoyl)piperazin-1-yl]-14,17-difluoro-6-(propan-2-yl)-11,12-dihydro-4H-1,18-(ethanediylidene)pyrido[4,3-e]pyrimido[1,6-g][1,4,7,9]benzodioxadiazacyclododecin-4-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Day, P.J, Cleasby, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-1084: An Orally Bioavailable and Low-Dose KRAS G12C Inhibitor.
J.Med.Chem., 67, 2024
|
|
8S8H
 
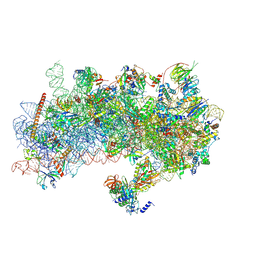 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8D
 
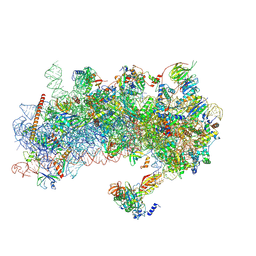 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8G
 
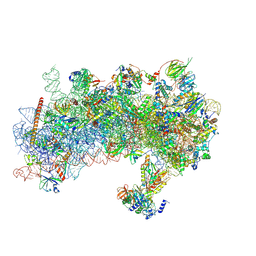 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-2.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8S8I
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 2024
|
|
8YKV
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8YKX
 
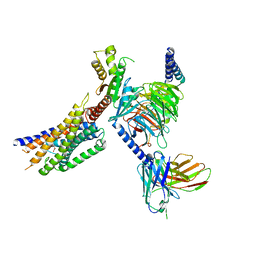 | | Cryo-EM structure of succinate receptor SUCR1 bound to maleic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
9AX6
 
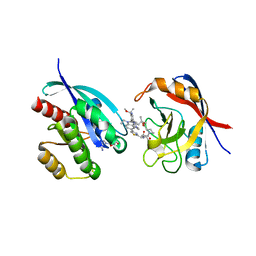 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
8YKW
 
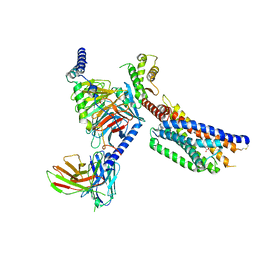 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
9AVL
 
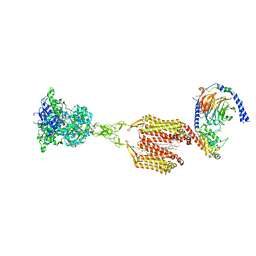 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
8YKA
 
 | | Cryo-EM structure of P97-VCPIP1 complex | | Descriptor: | Deubiquitinating protein VCPIP1, Transitional endoplasmic reticulum ATPase | | Authors: | Liu, Y, Lu, P, Gao, H, Li, F. | | Deposit date: | 2024-03-04 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Molecular Basis of VCPIP1 and P97/VCP Interaction Reveals Its Functions in Post-Mitotic Golgi Reassembly.
Adv Sci, 2024
|
|
9AVG
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AV7
 
 | |
9AV6
 
 | |
8S7J
 
 | | Coxsackievirus A9 bound with compound 20 (CL300) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-03-01 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 2024
|
|
