2MZD
 
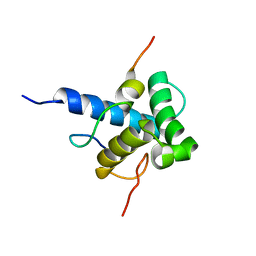 | | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Miller Jenkins, L.M, Feng, H, Durell, S.R, Tagad, H.D, Mazur, S.J, Tropea, J.E, Bai, Y, Appella, E. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the p300 Taz2-p53 TAD2 Complex and Comparison with the p300 Taz2-p53 TAD1 Complex.
Biochemistry, 54, 2015
|
|
3FXO
 
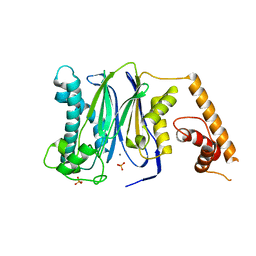 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 1 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
1GQ8
 
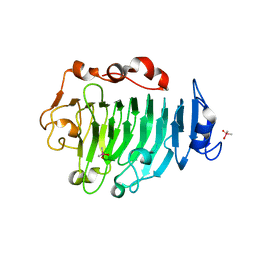 | | Pectin methylesterase from Carrot | | Descriptor: | CACODYLATE ION, PECTINESTERASE | | Authors: | Johansson, K, El-Ahmad, M, Friemann, R, Jornvall, H, Markovic, O, Eklund, H. | | Deposit date: | 2001-11-20 | | Release date: | 2002-04-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Plant Pectin Methylesterase
FEBS Lett., 514, 2002
|
|
2PYP
 
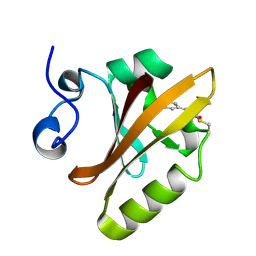 | | PHOTOACTIVE YELLOW PROTEIN, PHOTOSTATIONARY STATE, 50% GROUND STATE, 50% BLEACHED | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Borgstahl, G.E.O, Ng, K, Ren, Z, Pradervand, C, Burke, P, Srajer, V, Teng, T, Schildkamp, W, Mcree, D.E, Moffat, K, Getzoff, E.D. | | Deposit date: | 1997-02-03 | | Release date: | 1998-04-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a protein photocycle intermediate by millisecond time-resolved crystallography.
Science, 275, 1997
|
|
3FR4
 
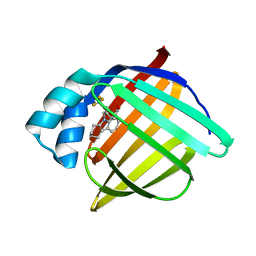 | | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte Fatty-Acid Binding Protein (A-FABP) inhibitors | | Descriptor: | 9-[2-(trifluoromethyl)benzyl]-2,3,4,9-tetrahydro-1H-carbazole-8-carboxylic acid, Fatty acid-binding protein, adipocyte | | Authors: | Barf, T, Hammer, K, Lehmann, F, Haile, S, Axen, E, Medina, C, Uppenberg, J, Svensson, S, Rondahl, L, Lundb ck, T. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | N-Benzyl-indolo carboxylic acids: Design and synthesis of potent and selective adipocyte fatty-acid binding protein (A-FABP) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3G6O
 
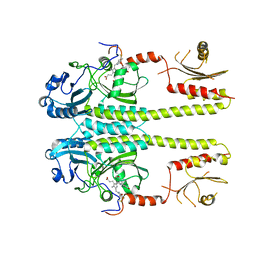 | |
1F5M
 
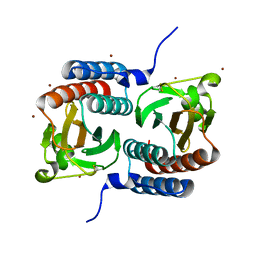 | | STRUCTURE OF THE GAF DOMAIN | | Descriptor: | BROMIDE ION, GAF | | Authors: | Ho, Y.S, Burden, L.M, Hurley, J.H. | | Deposit date: | 2000-06-15 | | Release date: | 2000-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor.
EMBO J., 19, 2000
|
|
3FXM
 
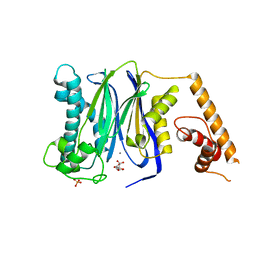 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 10 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
2FEX
 
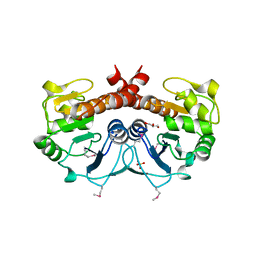 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
3BSO
 
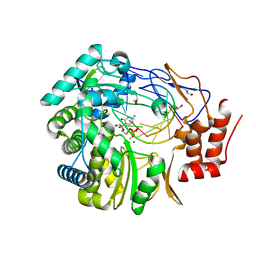 | |
2PG1
 
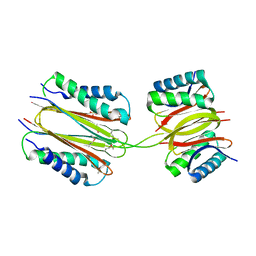 | |
2PHY
 
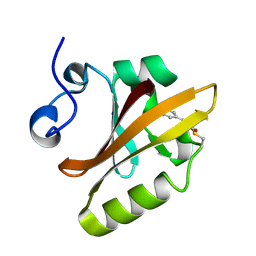 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Borgstahl, G.E.O, Getzoff, E.D. | | Deposit date: | 1995-04-12 | | Release date: | 1995-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore.
Biochemistry, 34, 1995
|
|
2FIX
 
 | |
2MZ8
 
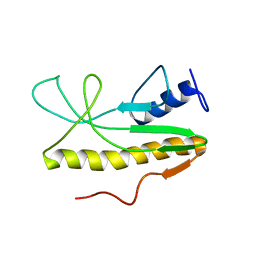 | | Solution NMR structure of Salmonella Typhimurium transcriptional regulator protein Crl | | Descriptor: | Sigma factor-binding protein Crl | | Authors: | Cavaliere, P, Levi-Acobas, F, Monteil, V, Bellalou, J, Mayer, C, Norel, F, Sizun, C. | | Deposit date: | 2015-02-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding interface between the Salmonella sigma (S)/RpoS subunit of RNA polymerase and Crl: hints from bacterial species lacking crl.
Sci Rep, 5, 2015
|
|
1ZTX
 
 | | West Nile Virus Envelope Protein DIII in complex with neutralizing E16 antibody Fab | | Descriptor: | Envelope protein, Heavy Chain of E16 Antibody, Light Chain of E16 Antibody | | Authors: | Nybakken, G.E, Oliphant, T, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of West Nile virus neutralization by a therapeutic antibody.
Nature, 437, 2005
|
|
1GV1
 
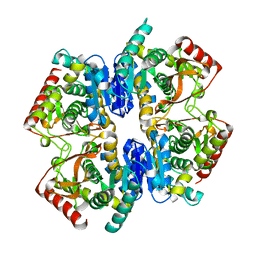 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
1ZUX
 
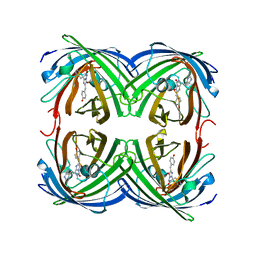 | | EosFP Fluorescent Protein- Green Form | | Descriptor: | green to red photoconvertible GPF-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1H3E
 
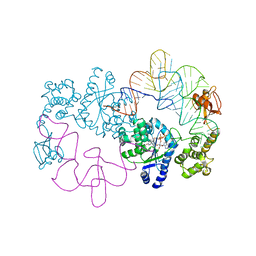 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with wild-type tRNAtyr(GUA) and with ATP and tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, ADENOSINE-5'-TRIPHOSPHATE, TYROSYL-TRNA SYNTHETASE, ... | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
2DW4
 
 | |
3C2W
 
 | |
2DII
 
 | | Solution structure of the BSD domain of human TFIIH basal transcription factor complex p62 subunit | | Descriptor: | TFIIH basal transcription factor complex p62 subunit | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Watabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the BSD domain of human TFIIH basal transcription factor complex p62 subunit
To be Published
|
|
2NDG
 
 | |
2DM5
 
 | | Thermodynamic Penalty Arising From Burial of a Ligand Polar Group Within a Hydrophobic Pocket of a Protein Receptor | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTANE-1,8-DIOL | | Authors: | Barratt, E, Bronowska, A, Vondrasek, J, Bingham, R, Phillips, S, Homans, S.W. | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic penalty arising from burial of a ligand polar group within a hydrophobic pocket of a protein receptor
J.Mol.Biol., 362, 2006
|
|
2DJ8
 
 | | Solution Structure of zf-MYND Domain of Protein CBFA2TI (Protein MTG8) | | Descriptor: | Protein CBFA2T1, ZINC ION | | Authors: | Niraula, T.N, Sasagawa, A, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of zf-MYND Domain of Protein CBFA2TI (Protein MTG8)
To be published
|
|
2DME
 
 | | Solution structure of the TFIIS domain II of human PHD finger protein 3 | | Descriptor: | PHD finger protein 3 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Watabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-21 | | Release date: | 2006-10-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TFIIS domain II of human PHD finger protein 3
To be Published
|
|
