7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
2NYE
 
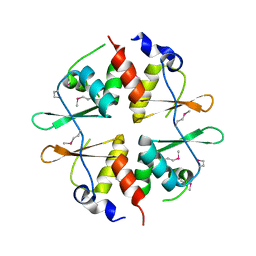 | | Crystal structure of the Bateman2 domain of yeast Snf4 | | Descriptor: | Nuclear protein SNF4 | | Authors: | Rudolph, M.J, Amodeo, G.A, Iram, S, Hong, S, Pirino, G, Carlson, M, Tong, L. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding.
Structure, 15, 2007
|
|
3B18
 
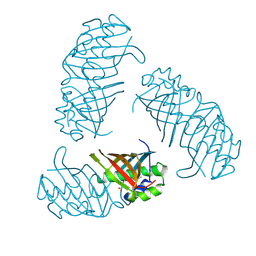 | |
2NYM
 
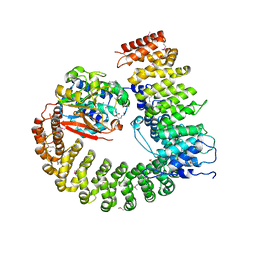 | | Crystal Structure of Protein Phosphatase 2A (PP2A) with C-terminus truncated catalytic subunit | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Chen, Y, Xing, Y, Xu, Y, Chao, Y, Lin, Z, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
5L9C
 
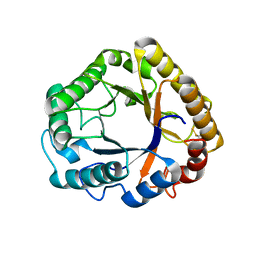 | | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Nemashkalov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinisyn, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose
to be published
|
|
5L8G
 
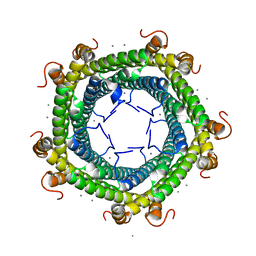 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5LB6
 
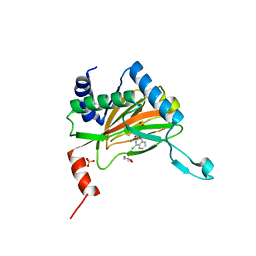 | |
3KCX
 
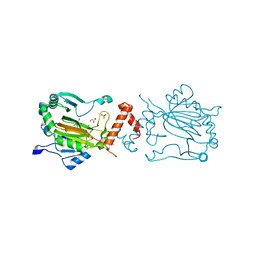 | | Factor inhibiting HIF-1 alpha in complex with Clioquinol | | Descriptor: | 5-chloro-7-iodoquinolin-8-ol, FE (II) ION, GLYCEROL, ... | | Authors: | Moon, H, Han, S, Choe, J. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human FIH-1 in complex with quinol family inhibitors
Mol.Cells, 29, 2010
|
|
5LBE
 
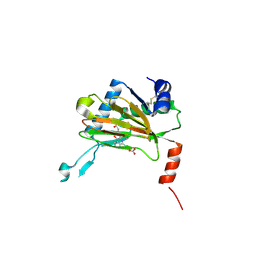 | |
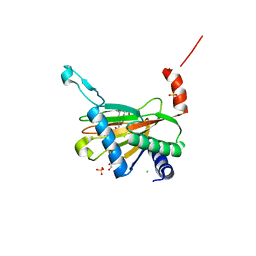
5LBC
 
 | |
3KD9
 
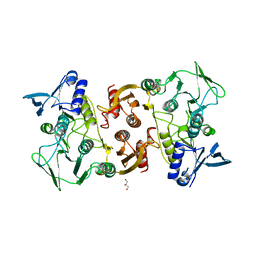 | |
7L6B
 
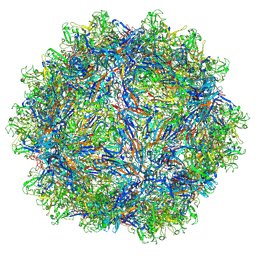 | | The empty AAV12 capsid | | Descriptor: | VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
2O9K
 
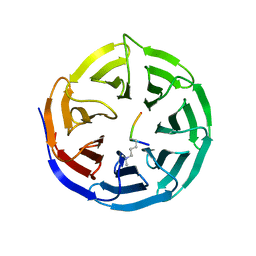 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
3B4R
 
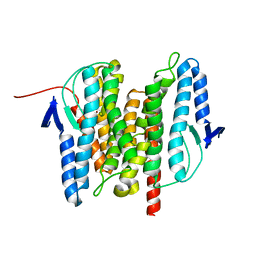 | | Site-2 Protease from Methanocaldococcus jannaschii | | Descriptor: | Putative zinc metalloprotease MJ0392, ZINC ION | | Authors: | Jeffrey, P.D, Feng, L, Yan, H, Wu, Z, Yan, N, Wang, Z, Shi, Y. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a site-2 protease family intramembrane metalloprotease.
Science, 318, 2007
|
|
5KJ8
 
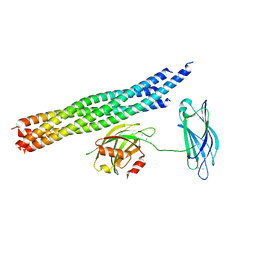 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from synchrotron diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
7DQC
 
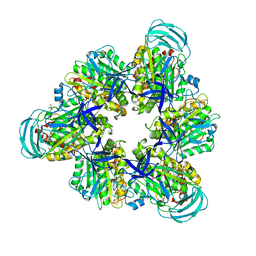 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
3KCF
 
 | |
3B5D
 
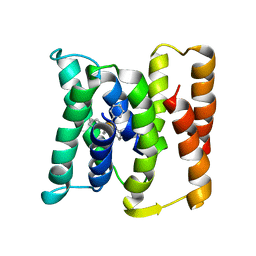 | |
2O4L
 
 | | Crystal Structure of HIV-1 Protease (Q7K, I50V) in Complex with Tipranavir | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
7KJ3
 
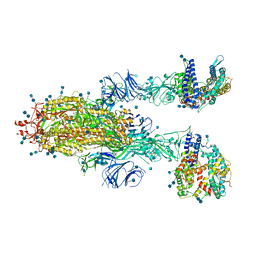 | | SARS-CoV-2 Spike Glycoprotein with two ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KNH
 
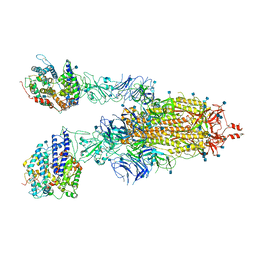 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
3KCY
 
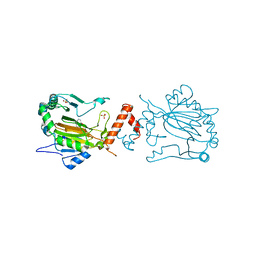 | | Factor inhibiting HIF-1 alpha in complex with 8-hydroxyquinoline | | Descriptor: | FE (II) ION, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Moon, H, Han, S, Choe, J. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of human FIH-1 in complex with quinol family inhibitors
Mol.Cells, 29, 2010
|
|
7DH1
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N2-Methylguanosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-12 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Specificity of GSDA Revealed by Cocrystal Structures and Binding Studies.
Int J Mol Sci, 23, 2022
|
|
2JU2
 
 | |
7D66
 
 | |
