4GHM
 
 | | Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Crystal Structure of the H233A mutant of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with preQ0
To be Published
|
|
4GIM
 
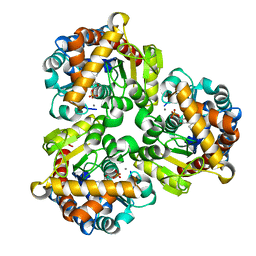 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Pseudouridine 5'-phosphate | | Descriptor: | MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, Pseudouridine-5'-phosphate glycosidase | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
4AD0
 
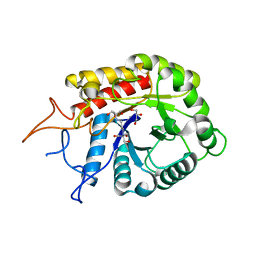 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2E9B
 
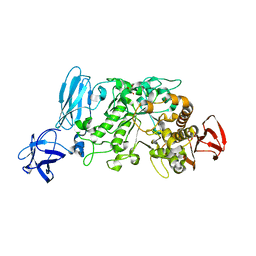 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with maltose | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
2YVB
 
 | |
2YVJ
 
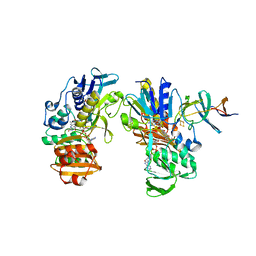 | | Crystal structure of the ferredoxin-ferredoxin reductase (BPHA3-BPHA4)complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2EA0
 
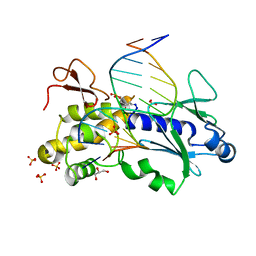 | | Crystal structure of the DNA repair enzyme endonuclease-VIII (Nei) from E. coli in complex with AP-site containing DNA substrate | | Descriptor: | 5'-D(P*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(P*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharov, D.O, Grollman, A.P, Shoham, G. | | Deposit date: | 2007-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active site plasticity of endonuclease VIII: Conformational changes compensating for different substrate and mutations of critical residues
To be Published
|
|
1Q5E
 
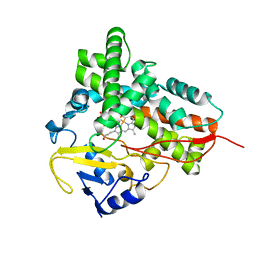 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
2YW6
 
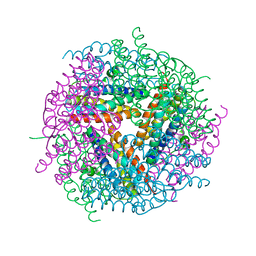 | | Structural studies of N terminal deletion mutant of Dps from Mycobacterium smegmatis | | Descriptor: | DNA protection during starvation protein | | Authors: | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-04-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
2Y64
 
 | | Xylopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2YBI
 
 | |
4AD4
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine and alpha-1,2- mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YAU
 
 | | X-ray structure of the Leishmania infantum tryopanothione reductase in complex with auranofin | | Descriptor: | 2,3,4,6-tetra-O-acetyl-1-thio-beta-D-glucopyranose, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G. | | Deposit date: | 2011-02-24 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Gold-Containing Drug Against Parasitic Polyamine Metabolism: The X-Ray Structure of Trypanothione Reductase from Leishmania Infantum in Complex with Auranofin Reveals a Dual Mechanism of Enzyme Inhibition.
Amino Acids, 42, 2012
|
|
4H1S
 
 | | Crystal Structure of a Truncated Soluble form of Human CD73 with Ecto-5'-Nucleotidase activity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, PHOSPHATE ION, ... | | Authors: | Heuts, D.P, Weissenborn, M.J, Olkhov, R.V, Shaw, A.M, Levy, C.W, Scrutton, N.S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a soluble form of human CD73 with ecto-5'-nucleotidase activity.
Chembiochem, 13, 2012
|
|
5NYG
 
 | |
4GQQ
 
 | | Human pancreatic alpha-amylase with bound ethyl caffeate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Williams, L.K, Brayer, G.D. | | Deposit date: | 2012-08-23 | | Release date: | 2012-10-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Order and disorder: differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target.
J.Med.Chem., 55, 2012
|
|
4GO0
 
 | |
1XGZ
 
 | | Structure of the N298S variant of human pancreatic alpha-amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, pancreatic, ... | | Authors: | Maurus, R, Begum, A, Kuo, H.H, Racaza, A, Numao, S, Overall, C.M, Withers, S.G, Brayer, G.D. | | Deposit date: | 2004-09-17 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase
PROTEIN SCI., 14, 2005
|
|
4AMW
 
 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-idosyl- fluoride | | Descriptor: | 5-fluoro-alpha-L-idopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1PQA
 
 | | Trypsin with PMSF at atomic resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
2ZMA
 
 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
2E8Z
 
 | | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 complexed with alpha-cyclodextrin | | Descriptor: | ACETATE ION, AmyX protein, CALCIUM ION, ... | | Authors: | Mikami, B, Malle, D, Utsumi, S, Iwamoto, H, Katsuya, Y. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of pullulanase type I from Bacillus subtilis str. 168 in complex with maltose and alpha-cyclodextrin
To be Published
|
|
4GVH
 
 | |
2EII
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-valine and NAD. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD.
To be Published
|
|
2ZR0
 
 | | MSRECA-Q196E mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
