4K7W
 
 | | Crystal structure of Zn3-hUb(human ubiquitin) adduct from a solution 100 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ZINC ION, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
4KQ6
 
 | | Product complex of lumazine synthase from candida glabrata | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 6,7-dimethyl-8-ribityllumazine synthase, GLYCEROL, ... | | Authors: | Shankar, M, Wilbanks, S.M, Nakatani, Y, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Catalysis product captured in lumazine synthase from the fungal pathogen Candida glabrata.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2DI0
 
 | | Solution Structure of the CUE Domain in the Human Activating Signal Cointegrator 1 Complex Subunit 2 (ASCC2) | | Descriptor: | Activating signal cointegrator 1 complex subunit 2 | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the CUE Domain in the Human Activating Signal Cointegrator 1 Complex Subunit 2 (ASCC2)
To be Published
|
|
3VCY
 
 | | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase), from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Bensen, D.C, Rodriguez, S, Nix, J, Cunningham, M.L, Tari, L.W. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3RM9
 
 | | AMCase in complex with Compound 3 | | Descriptor: | 4-(4-chlorophenyl)piperazine-1-carboximidamide, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3ARW
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with chelerythrine | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
1VST
 
 | | Symmetric Sulfolobus solfataricus uracil phosphoribosyltransferase with bound PRPP and GTP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kadziola, A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and kinetic studies of the allosteric transition in Sulfolobus solfataricus uracil phosphoribosyltransferase: Permanent activation by engineering of the C-terminus
J.Mol.Biol., 393, 2009
|
|
3RNK
 
 | | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain | | Descriptor: | Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain
To be Published
|
|
2DMW
 
 | | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1 | | Descriptor: | synaptobrevin-like 1 variant | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1
To be Published
|
|
1IWO
 
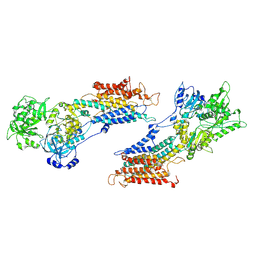 | |
3VW8
 
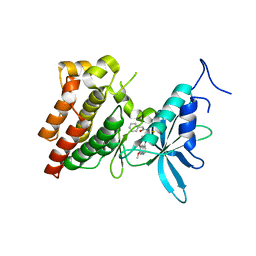 | | Crystal structure of human c-Met kinase domain with its inhibitor | | Descriptor: | CHLORIDE ION, Hepatocyte growth factor receptor, N-({4-[(6,7-dimethoxyquinolin-4-yl)oxy]phenyl}carbamothioyl)-2-phenylacetamide | | Authors: | Matsumoto, S, Miyamoto, N, Hirayama, T, Oki, H, Okada, K, Tawada, M, Iwata, H, Miki, H, Nakamura, K, Hori, A, Imamura, S. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design, synthesis, and evaluation of imidazo[1,2-b]pyridazine and imidazo[1,2-a]pyridine derivatives as novel dual c-Met and VEGFR2 kinase inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
3VZL
 
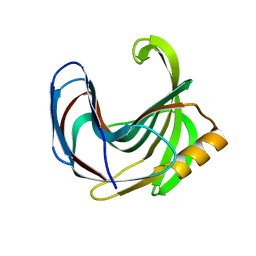 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
4KEZ
 
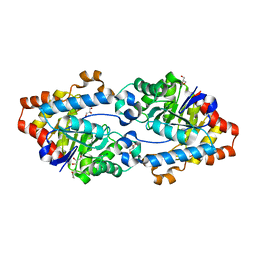 | | Crystal structure of SsoPox W263F | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
1VR8
 
 | |
2CT5
 
 | | Solution Structure of the zinc finger BED domain of the zinc finger BED domain containing protein 1 | | Descriptor: | ZINC ION, Zinc finger BED domain containing protein 1 | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the zinc finger BED domain of the zinc finger BED domain containing protein 1
To be Published
|
|
4KHT
 
 | | Triple helix bundle of GP41 complexed with fab 8066 | | Descriptor: | 8066 heavy chain, 8066 light chain, Gp41 helix | | Authors: | Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2013-05-01 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Complexes of neutralizing and non-neutralizing affinity matured Fabs with a mimetic of the internal trimeric coiled-coil of HIV-1 gp41.
Plos One, 8, 2013
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4KDX
 
 | | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group p21, form(1) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase domain | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group P21, form(1)
To be Published
|
|
2CXK
 
 | |
3VAK
 
 | | Structure of U2AF65 variant with BrU5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
1E56
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural substrate DIMBOA-beta-D-glucoside | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase- Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2CT0
 
 | | Solution structure of the RING domain of the Non-SMC element 1 protein | | Descriptor: | Non-SMC element 1 homolog, ZINC ION | | Authors: | Miyamoto, K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the Non-SMC element 1 protein
To be Published
|
|
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FAI
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-2M and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2S,5S,9R)-5-(HYDROXYMETHYL)-8,9-DIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | Authors: | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
