7XKP
 
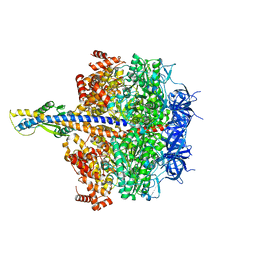 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7CGR
 
 | |
5N5L
 
 | |
5NDT
 
 | |
8DXE
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-amino-6-fluorobenzonitrile at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-6-fluorobenzonitrile, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
3EAO
 
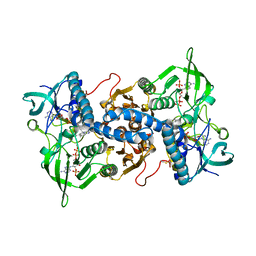 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with oxidized C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
4S2K
 
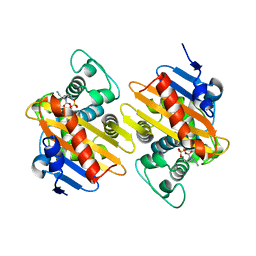 | | OXA-48 in complex with Avibactam at pH 7.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
6BLR
 
 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E6SS | | Descriptor: | 1,2-ETHANEDIOL, H2-Ab1 protein, IAg7 alpha chain | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9PW7
 
 | | Myeloid cell leukemia-1 (Mcl-1) complexed with compound 13 | | Descriptor: | (2S,4R,5S,12P,23R)-11-chloro-7-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-27,28-dimethoxy-4,15-dimethyl-32-oxo-19-oxa-2,5,15,16,23-pentaazaheptacyclo[21.6.1.1~2,6~.1~5,8~.0~12,31~.0~13,17~.0~26,30~]dotriaconta-1(30),6,8(31),9,11,13,16,24,26,28-decaene-24-carboxylic acid (non-preferred name), Maltodextrin-binding protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhao, B, Fesik, S.W. | | Deposit date: | 2025-08-04 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Macrocyclic Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors that Demonstrate Potent Cellular Efficacy and In Vivo Activity in a Mouse Solid Tumor Xenograft Model.
J.Med.Chem., 68, 2025
|
|
7KXC
 
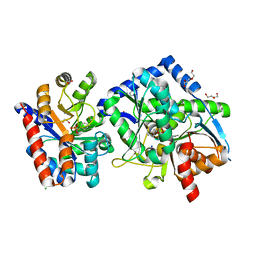 | | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The aminoacrylate form of the wild-type Salmonella typhimurium Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, sodium ion at the metal coordination site and benzimidazole (BZI) at the enzyme beta-site at 1.30 Angstrom resolution. One of the beta-Q114 rotamer conformations allows a hydrogen bond to form with the PLP oxygen at the position 3 in the ring.
To be Published
|
|
3DK8
 
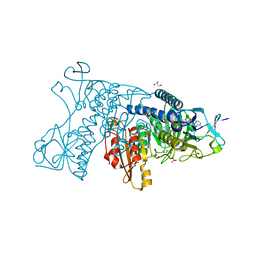 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
7KWV
 
 | |
4XZH
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0048 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-chloro-3-nitrobenzyl)-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZI
 
 | | Crystal structure of human Aldose Reductase complexed with NADP+ and JF0049 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,4-dioxo-3-(2,3,4,5-tetrabromo-6-methoxybenzyl)-3,4-dihydropyrimidin-1(2H)-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
7LGX
 
 | | The aminoacrylate form of mutant beta-K167T Salmonella typhimurium Tryptophan Synthase in complex with with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, benzimidazole (BZI) at the enzyme beta site and cesium ion at the metal coordination site at 1.80 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-01-21 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aminoacrylate form of mutant beta-K167T Salmonella typhimurium Tryptophan Synthase in complex with with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, benzimidazole (BZI) at the enzyme beta site and cesium ion at the metal coordination site at 1.80 Angstrom resolution.
To be Published
|
|
9DRA
 
 | | Crystal structure of Catechol 1,2-dioxygenase from Burkholderia multivorans (Iron and 4,5-dichloro-1,2-catechol bound) | | Descriptor: | (6R)-3,4-dichloro-6-hydroxycyclohex-3-en-1-one, CALCIUM ION, Catechol 1,2-dioxygenase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-09-25 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Catechol 1,2-dioxygenase from Burkholderia multivorans (Iron and 4,5-dichloro-1,2-catechol bound)
To be published
|
|
3U7Z
 
 | |
9D6W
 
 | |
9DL7
 
 | | Structure of proline utilization A complexed with 1-(4-fluorophenyl)thiourea | | Descriptor: | 1-(4-fluorophenyl)thiourea, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2024-09-10 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic Fragment Screening of a Bifunctional Proline Catabolic Enzyme Reveals New Inhibitor Templates for Proline Dehydrogenase and L-Glutamate-gamma-semialdehyde Dehydrogenase.
Molecules, 29, 2024
|
|
5N4U
 
 | | Crystal structure of human Pim-1 kinase in complex with a consensuspeptide and fragment like molekule 5-(2-amino-1,3-thiazol-4-yl)-1,3-dihydrobenzimidazol-2-one | | Descriptor: | 5-(2-azanyl-1,3-thiazol-4-yl)-1,3-dihydrobenzimidazol-2-one, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
8CRK
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
9D7B
 
 | | OXA-58-NA-1-157 7.5 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
9D79
 
 | | OXA-58-NA-1-157 1.5 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
9D7D
 
 | | OXA-58-NA-1-157 20 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase, ... | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
8R4X
 
 | | Structure of Chitinase-3-like protein 1 in complex with inhibitor 30 | | Descriptor: | (2~{S},5~{S})-4-[1-(4-chloranylpyridin-2-yl)piperidin-4-yl]-5-[(4-chlorophenyl)methyl]-2-methyl-morpholine, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
