6VNG
 
 | | JAK2 JH1 in complex with PN2-118 | | Descriptor: | N-{2-fluoro-5-[(2-{[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]phenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNH
 
 | | JAK2 JH1 in complex with PN2-123 | | Descriptor: | N-{5-[(2-{[3,5-difluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]-2-fluorophenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNF
 
 | | JAK2 JH1 in complex with MA9-086 | | Descriptor: | N~4~-[1-(tert-butylsulfonyl)-2,3-dihydro-1H-indol-6-yl]-N~2~-[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]-5-methylpyrimidine-2,4-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VN8
 
 | | JAK2 JH1 in complex with baricitinib | | Descriptor: | Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNJ
 
 | | JAK2 JH1 in complex with PN4-014 | | Descriptor: | 3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNM
 
 | | JAK2 JH1 in complex with SY5-103 | | Descriptor: | 4-[1-(but-3-en-1-yl)-1H-pyrazol-4-yl]-N-[4-(piperidin-4-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-2-amine, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNI
 
 | | JAK2 JH1 in complex with PN3-115 | | Descriptor: | 2-{5-[(2-{[3,5-difluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]-2-fluorophenyl}-1lambda~6~,2-thiazolidine-1,1-dione, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNL
 
 | | JAK2 JH1 in complex with SG3-179 | | Descriptor: | 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNK
 
 | | JAK2 JH1 in complex with PN4-073 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VS3
 
 | | JAK2 JH1 in complex with BL2-057 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VSN
 
 | | JAK2 JH1 in complex with BL2-110 | | Descriptor: | (3S)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-02-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6JZ5
 
 | |
6JZ8
 
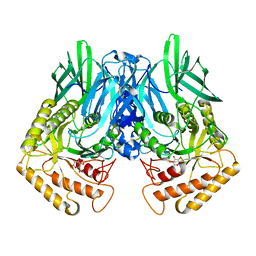 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6W4Z
 
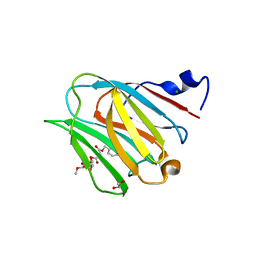 | | Galectin-8N terminal domain in complex with Methyl 3-O-[3-O-benzyloxy]-malonyl-beta-D-galactopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galectin-8, ... | | Authors: | Patel, B, Kishor, C, Blanchard, H. | | Deposit date: | 2020-03-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rational Design and Synthesis of Methyl-beta-d-galactomalonyl Phenyl Esters as Potent Galectin-8 N Antagonists.
J.Med.Chem., 63, 2020
|
|
1MZC
 
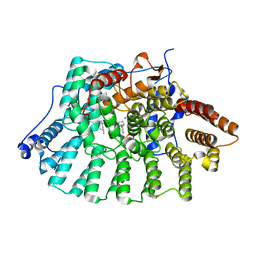 | | Co-Crystal Structure Of Human Farnesyltransferase With Farnesyldiphosphate and Inhibitor Compound 33a | | Descriptor: | 2-[3-(3-ETHYL-1-METHYL-2-OXO-AZEPAN-3-YL)-PHENOXY]-4-[1-AMINO-1-(1-METHYL-1H-IMIDIZOL-5-YL)-ETHYL]-BENZONITRILE, FARNESYL DIPHOSPHATE, Protein Farnesyltransferase alpha Subunit, ... | | Authors: | deSolms, S.J, Ciccarone, T.M, MacTough, S.C, Shaw, A.W, Buser, C.A, Ellis-Hutchings, M, Fernandes, C, Hamilton, K.A, Huber, H.E, Kohl, N.E, Lobell, R.B, Robinson, R.G, Tsou, N.N, Walsh, E.S, Graham, S.L, Beese, L.S, Taylor, J.S. | | Deposit date: | 2002-10-07 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual Protein Farnesyltransferase-Geranylgeranyltransferase-I Inhibitors as Potential Cancer Chemotherapeutic Agents.
J.Med.Chem., 46, 2003
|
|
8QQ4
 
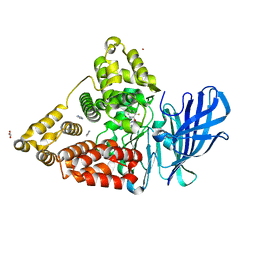 | | LTA4 hydrolase in complex with compound 6(R) | | Descriptor: | (2R)-2-azanyl-3-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-10-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
8QPN
 
 | | LTA4 hydrolase in complex with compound 6(S) | | Descriptor: | (2S)-2-azanyl-3-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
8QOW
 
 | | LTA4 hydrolase in complex with compound 2(S) | | Descriptor: | (2~{S})-2-azanyl-3-[3-[4-[3-fluoranyl-5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxyphenyl]pyrazol-1-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
3W4I
 
 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
7MR9
 
 | |
7MRA
 
 | |
7MRB
 
 | |
7PFY
 
 | |
7PG1
 
 | |
