4IJL
 
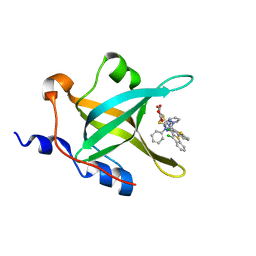 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, {[5-(3-chloro-1-benzothiophen-2-yl)-4-phenyl-4H-1,2,4-triazol-3-yl]sulfanyl}acetic acid | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
2Q7K
 
 | |
2Q7L
 
 | |
2Q7J
 
 | |
3G6J
 
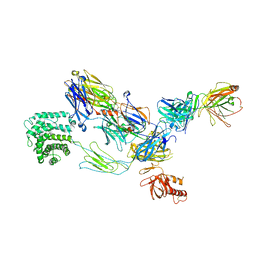 | | C3b in complex with a C3b specific Fab | | Descriptor: | CALCIUM ION, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Wiesmann, C. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement
J.Biol.Chem., 284, 2009
|
|
2ORU
 
 | |
2RIS
 
 | |
2PNX
 
 | |
2Q7I
 
 | |
2RIU
 
 | | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase- alternate interpreation | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Stenkamp, R.E, Le Trong, I. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6BSM
 
 | | BMP1 complexed with a reverse hydroxamate - compound 4 | | Descriptor: | Bone morphogenetic protein 1, N-({[(2R)-2-{[hydroxy(hydroxymethyl)amino]methyl}heptanoyl]amino}methyl)-7-methoxy-1-benzofuran-2-carboxamide, ZINC ION | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTO
 
 | | BMP1 complexed with (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid | | Descriptor: | (2~{S})-2-[[(1~{R},3~{S},4~{S})-2-[(2~{R})-2-[2-(oxidanylamino)-2-oxidanylidene-ethyl]heptanoyl]-2-azabicyclo[2.2.1]heptan-3-yl]carbonylamino]-2-phenyl-ethanoic acid, 1,2-ETHANEDIOL, Bone morphogenetic protein 1, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BSL
 
 | | BMP1 complexed with a reverse hydroxymate - compound 22 | | Descriptor: | 1,2-ETHANEDIOL, Bone morphogenetic protein 1, N-(2-ethoxy-4-{5-[({[(2R)-2-{(1R)-1-[formyl(hydroxy)amino]propyl}heptanoyl]amino}methyl)carbamoyl]furan-2-yl}benzene-1-carbonyl)-L-aspartic acid, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTN
 
 | | BMP1 complexed with a reverse hydroxymate - compound 1 | | Descriptor: | (3S)-6-cyclohexyl-3-{3-[(dimethylamino)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyhexanamide, 1,2-ETHANEDIOL, Bone morphogenetic protein 1, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTQ
 
 | | BMP1 complexed with a hydroxamate - compound 2 | | Descriptor: | 1,2-ETHANEDIOL, Bone morphogenetic protein 1, N~2~-[(2H-1,3-benzodioxol-5-yl)methyl]-N-hydroxy-N~2~-[(4-methoxyphenyl)sulfonyl]-3-thiophen-2-yl-D-alaninamide, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
6BTP
 
 | | BMP1 complexed with a hydroxamate | | Descriptor: | (1R,3S,4S)-2-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]heptanoyl}-N-(3-methoxypyrazin-2-yl)-2-azabicyclo[2.2.1]heptane-3-carboxamide, Bone morphogenetic protein 1, THIOCYANATE ION, ... | | Authors: | Gampe, R, Shewchuk, L. | | Deposit date: | 2017-12-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Reverse Hydroxamate Inhibitors of Bone Morphogenetic Protein 1.
ACS Med Chem Lett, 9, 2018
|
|
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
5UBI
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni with bound PRPP | | Descriptor: | 1,2-ETHANEDIOL, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
5UBH
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni with bound ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
5UBG
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni with bound Phosphoribosyl-ATP | | Descriptor: | ATP phosphoribosyltransferase, CHLORIDE ION, PHOSPHORIBOSYL ATP, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
5UB9
 
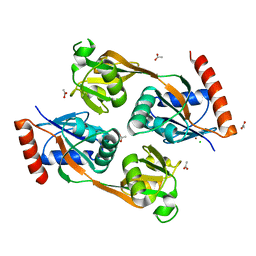 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni | | Descriptor: | ACETATE ION, ATP phosphoribosyltransferase, CHLORIDE ION, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
6NPE
 
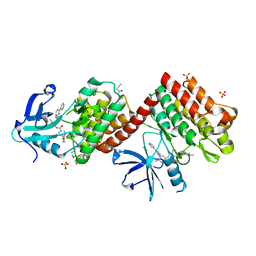 | | C-abl Kinase domain with the activator(cmpd6), 2-cyano-N-(4-(3,4-dichlorophenyl)thiazol-2-yl)acetamide | | Descriptor: | 2-cyano-~{N}-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]ethanamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, NONAETHYLENE GLYCOL, ... | | Authors: | campobasso, N. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and Optimization of Novel Small c-Abl Kinase Activators Using Fragment and HTS Methodologies.
J. Med. Chem., 62, 2019
|
|
6NPU
 
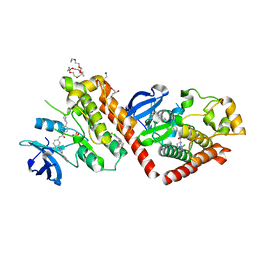 | |
6NPV
 
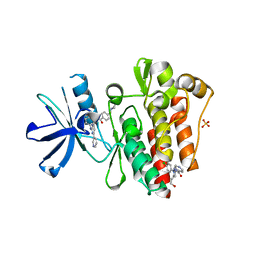 | |
5TCY
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
